Method for identifying DNA enhancer element based on sequence frequency information
A technology of frequency information and sequence information, which is applied in the field of functional component prediction algorithms to achieve high prediction accuracy and good prediction performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
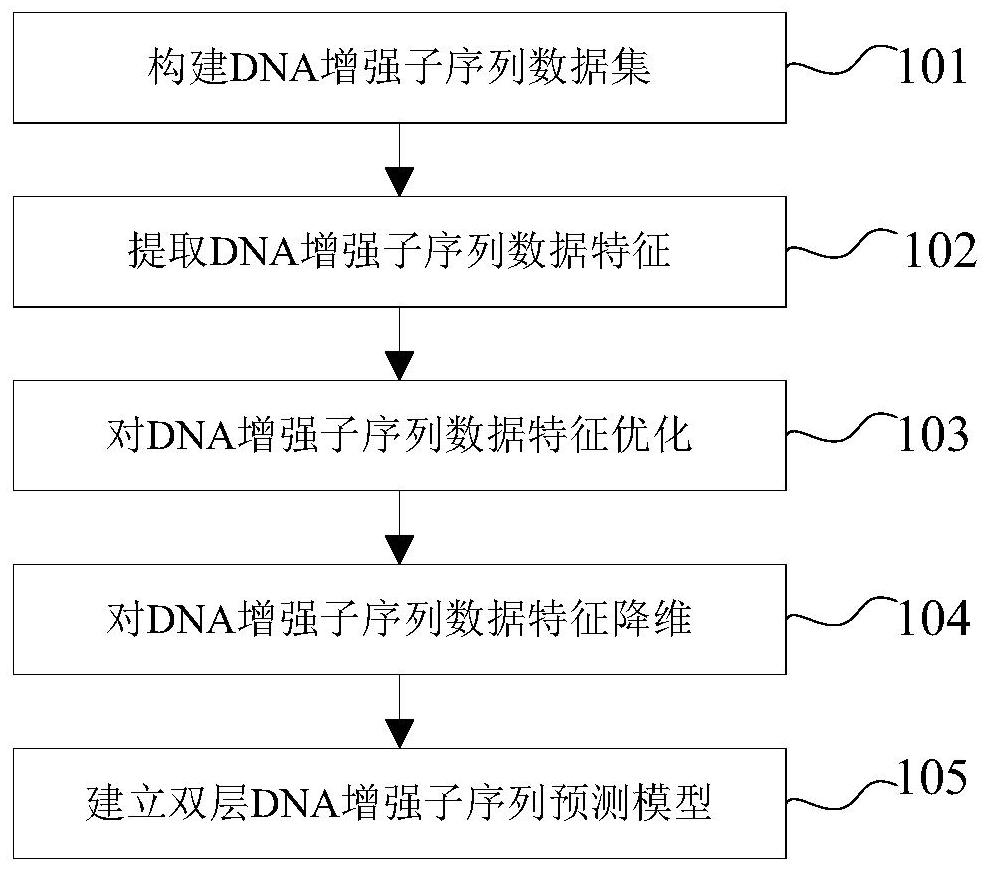
[0039] The present invention is described in detail below in conjunction with accompanying drawing
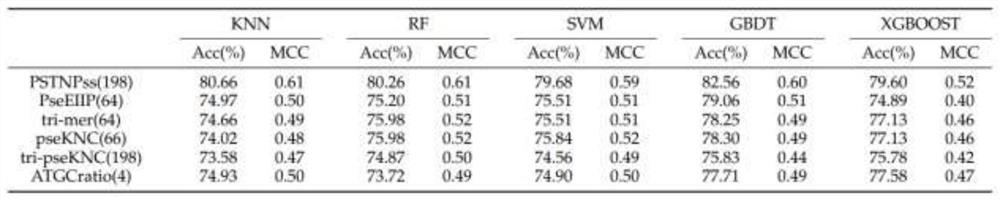
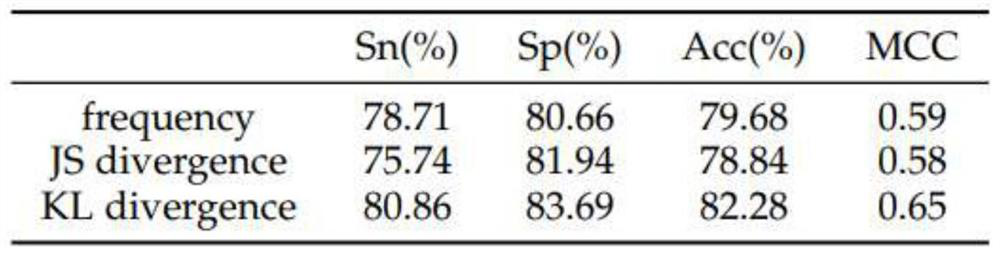
[0040] The enhancer involved in the present invention is a short DNA fragment, which can regulate the transcription level by recruiting transcription factors, forming a transcription complex and binding to the promoter site during the transcription process. By predicting the enhancers in the DNA sequence, it can help researchers in the biological field to find the reasons for the abnormal increase of transcription levels, and the enhancers of different strengths make it possible to program and regulate the transcription level. At present, the identification of enhancers mainly relies on biological experiments, but the experimental methods are often time-consuming and laborious; in contrast, it is easier and faster to use machine learning methods to predict enhancers.
[0041] The basic idea of the invention is to extract the position-specific information of the enhancer seque...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com