Method for identifying DNA promoter element based on information theory
A promoter and information theory technology, applied in the field of identifying DNA promoter elements based on information theory, can solve the problems of time-consuming and material-consuming, and achieve good prediction performance and high prediction accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
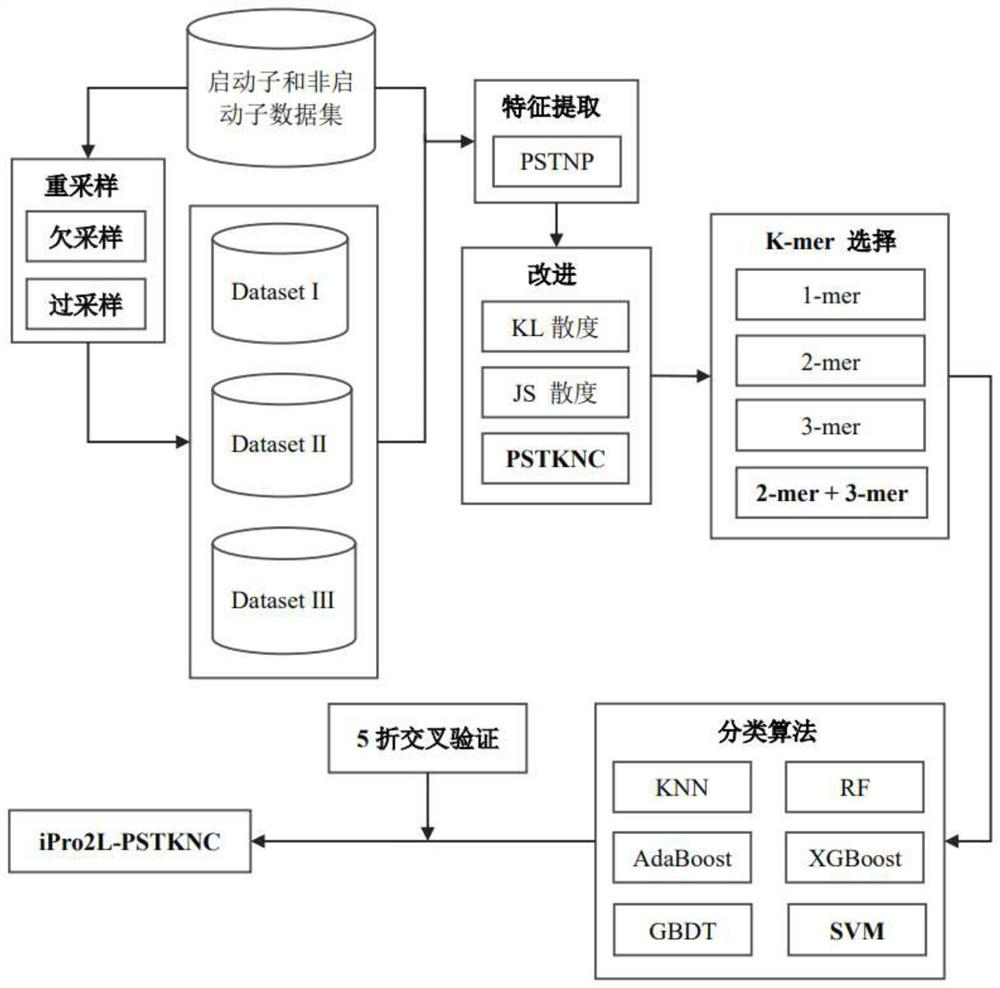
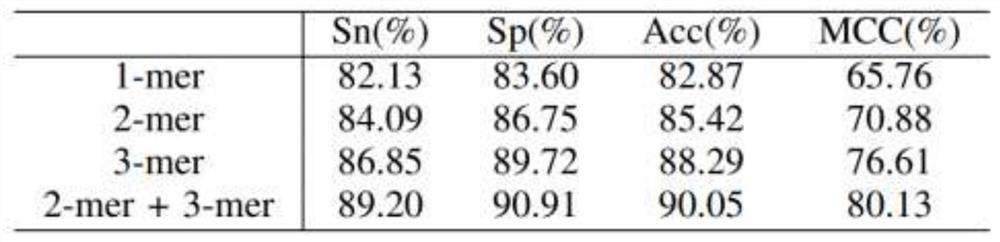
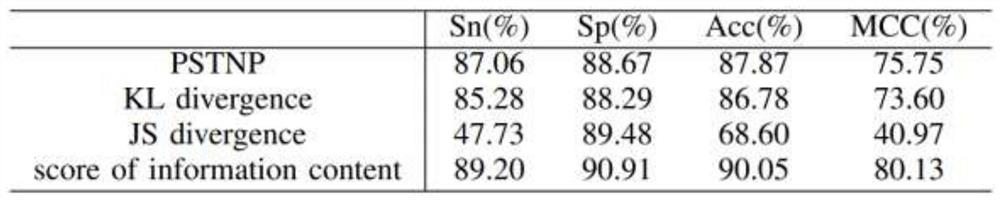
[0042] Promoter determines the initiation of DNA sequence-specific transcription and is an important regulatory element necessary for gene expression. Identifying and locating promoters is helpful for the accurate positioning of genes, and will play an important guiding role in annotating the structure and function information of biological genomes. In the process of gene transcription, when RNA polymerase specifically binds to a specific promoter, it needs a specific σ protein factor to assist in the recognition. Therefore, the σ factor is often used to mark the type of promoter, which is recorded as σ 24 , σ 28 , σ 32 , σ 38 , σ 54 , σ 70 . At present, the traditional biological experimental methods used to identify promoters and their types are time-consuming, laborious and costly. In contrast, using bioinformatics algorithms to identify and classify is a more economical and convenient method.
[0043] The basic idea of the invention is to extract the position-speci...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com