A method and application for predicting homologous recombination deficiency in ovarian cancer based on genomic copy number variation biomarkers
A biomarker and copy number variation technology, applied in the field of biomedicine, achieves good accuracy, improves survival rate, and predicts stable effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Application of a method for predicting homologous recombination deficiency in ovarian cancer based on copy number variation
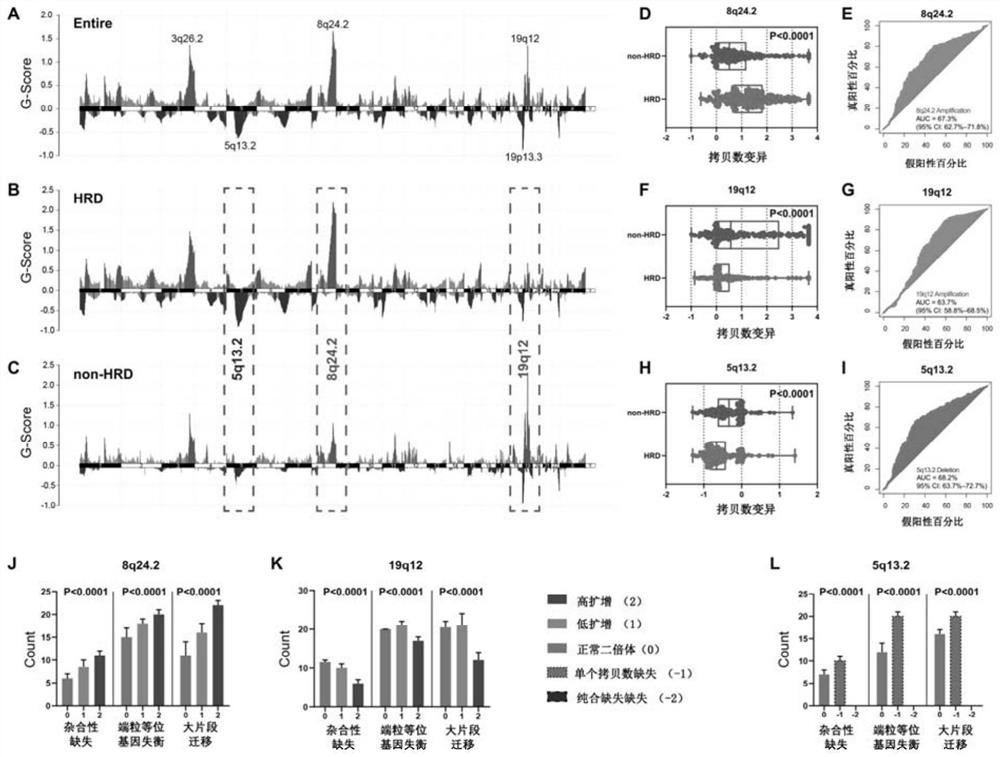
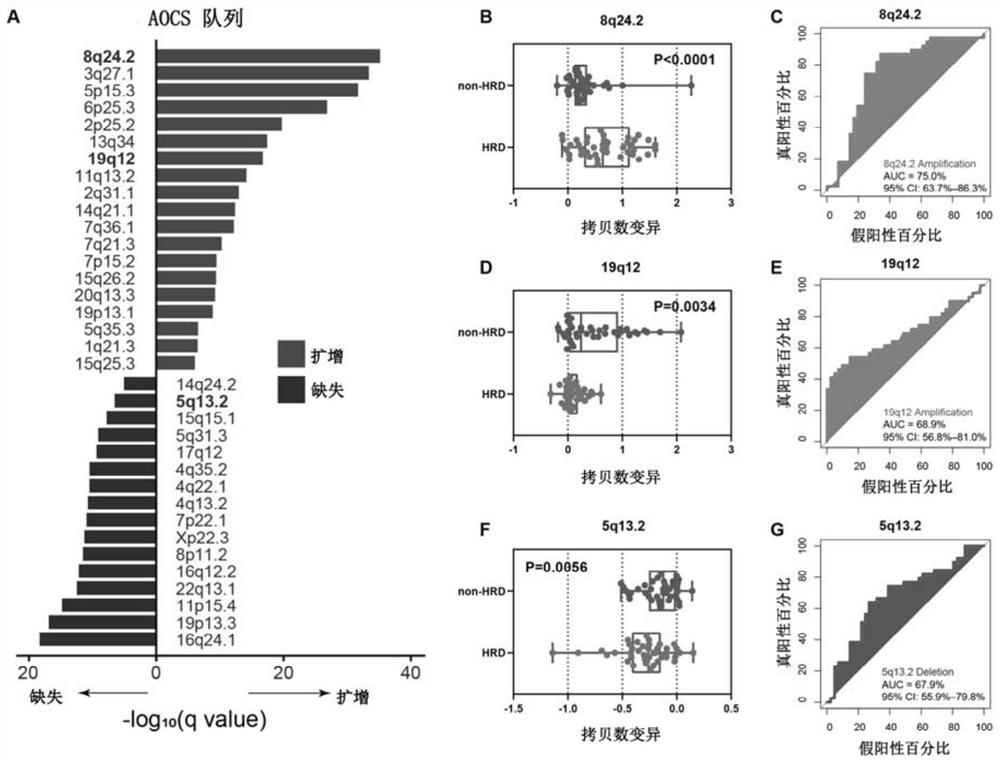
[0035] The flow process of the present invention research is as Figure 5 As shown, in Example 1, 587 ovarian cancer patients from the TCGA-OV cohort were included in the study, and their clinical characteristics and DNA sequencing data were obtained from https: / / www.cancer.gov / tcga.
[0036] According to HRD-score (HRD score), 587 patients were divided into HRD group (HRD score ≥ 42) and non-HRD group (HRD score < 42), HRD score is loss of heterozygosity (Loss of heterozygosis, LOH), large Unweighted sum of Large-scale state Transition (LST) and Telomeric Allelic Imbalance (TAI).
[0037] GISTIC 2.0 was used to measure CNV profiles at the subchromosomal level of the tumor genome, and the segment files of the TCGA-OV cohort (recording the copy number of each locus in the genome) were used as input to GSITIC 2.0 to quantify the CNV status of chro...
Embodiment 2
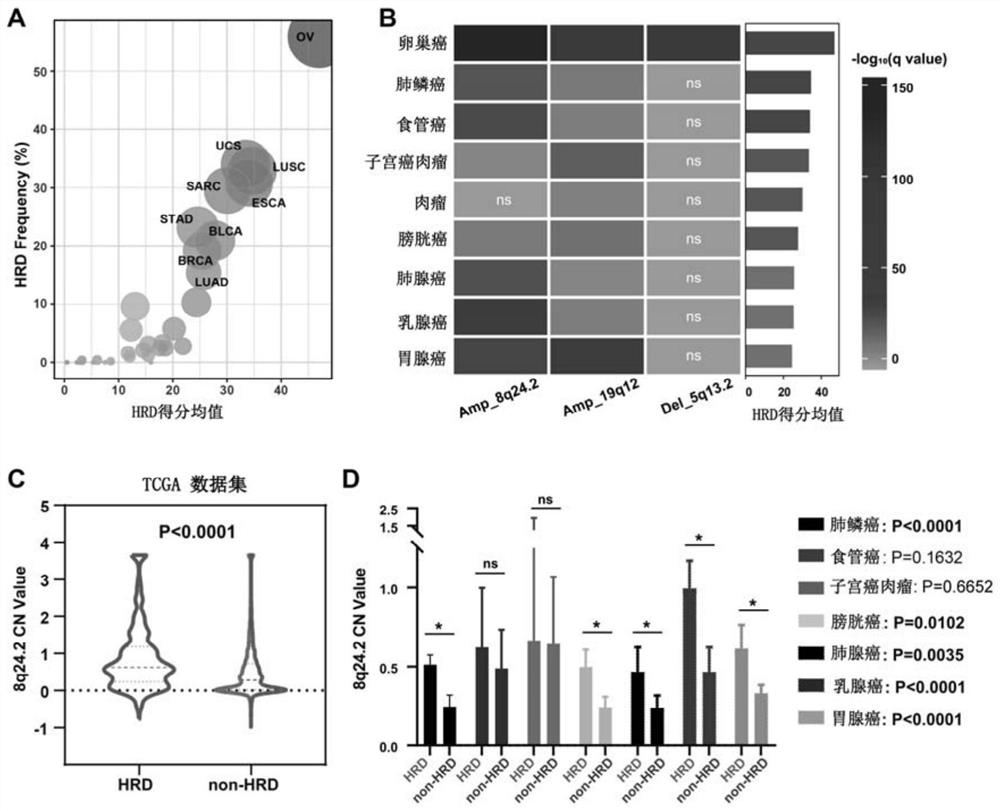
[0047] Application of a copy number variation-based biomarker to predict pan-cancer homologous recombination deficiency
[0048] The DNA sequencing results of 10,635 patients with 33 cancer types from the TCGA pan-cancer cohort were analyzed to extend the predictive role of CNV for homologous recombination defects in ovarian cancer to other cancer types.
[0049] Calculate the average HRD score for each tumor to obtain the distribution of homologous recombination deficiency status in different tumors, found ovarian serous cystadenocarcinoma (OV), uterine carcinosarcoma (UCS), lung squamous cell carcinoma (LUSC), sarcoma (SARC) , Patients with esophageal cancer (ESCA). HRD scores were predominantly higher in patients with gastric adenocarcinoma (STAD), bladderurethral carcinoma (BLCA), breast cancer (BRCA), and lung adenocarcinoma (LUAD), whereas patients with thyroid cancer (THCA) and acute myelogenous leukemia (AML) had HRD scores lowest( image 3 A). This is consistent wi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com