Method for improving homologous recombination efficiency based on CRISPR gene editing system
A gene editing and homologous recombination technology, applied in the field of genetic engineering, can solve problems such as potential safety hazards, decreased ability of cells to resist damage, and low homologous recombination rate, achieving high safety and reducing the difficulty of screening
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 Biotin-streptavidin strong interaction improves homologous recombination efficiency
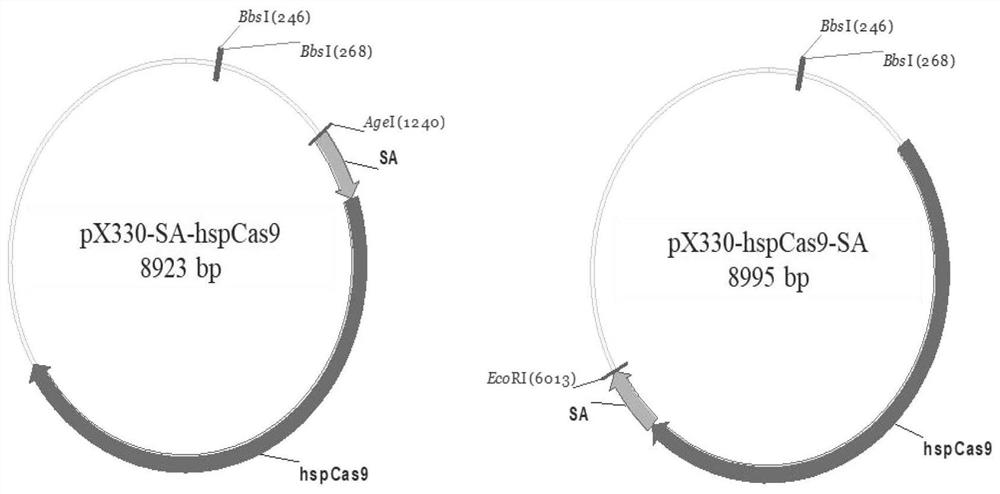
[0040] 1. Construction of Cas9 plasmid integrated with streptavidin and constructs of PX330-HSPCas9-SA plasmid containing GRNA
[0041] (1) Construction of Cas9 plasmid integrating streptavidin
[0042] The chain enzyme affinity is contained in PX330-HSPCAS9 with a PX330-HSPCAS9 plasmid of PX330-HSPCAS9.
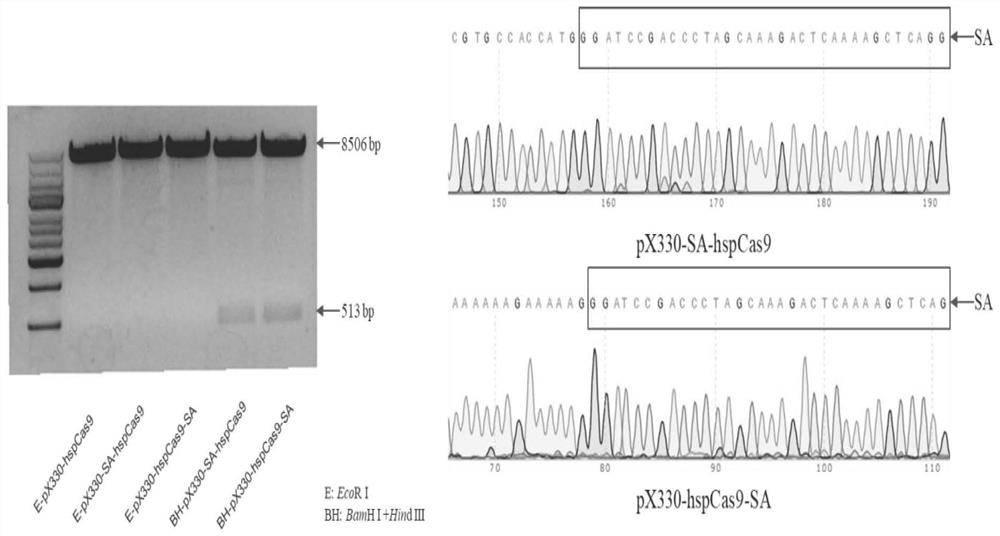
[0043] Review the sequence of streptavidin (SA) sequences in Synchronization OfsecretoryProtein Traffic In Population; Nature Methods. Chain enzyme affinity fragment. The synthesized streptavidin fragment was used for PCR amplification, and the amplification primer was HSPCas9-SA-F / HspCas9-Sa-R, and the nucleotide sequence such as SEQ ID NO: 4 / SEQ ID NO: 5 showed The sequence information of the PX330-HSPCas9 skeleton skeleton skeleton skeleton cut position is reserved.
[0044] The PX330-HSPCAS9 skeleton is separately digested with AgeI and EcoRI, and streptavidin is fused ...
Embodiment 2
[0076] Example 2 Biotin-streptavidin strong interaction with NHEJ repaired red green carrier indirect screening homologous recombination combined with establishing an efficient homologous recombination method
[0077] 1. Construction of Cas9 plasmid integrated with streptavidin
[0078] The build step is the same as that of the first step (1) of Example 1.
[0079] 2, the corresponding homologous donor of biotin modified homogeneous donor is constructed
[0080] (1) Construction of the original homologous donor template (Bio-egfp-donor):
[0081] The build step is the same as the step (1) step (1) of Example 1.
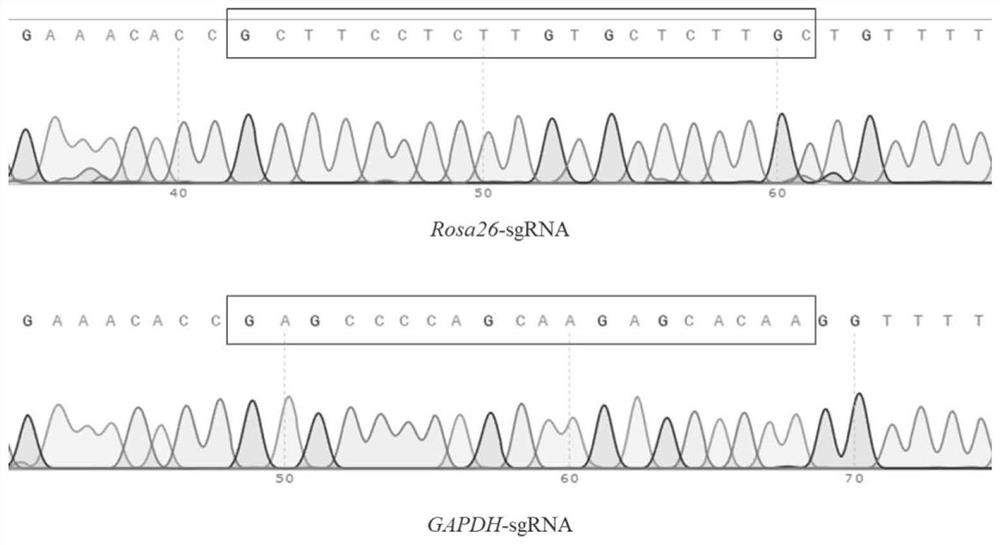
[0082] (2) Construction of the complete SGRNA plasmid (PX330-GapdhsGRNA-HSPCAS9) containing the U6 promoter and the terminator sequence: Based on the PX330-HSPCAS9 plasmid, ROSA26-GRNA or GAPDH-SGRNA is connected to BBSIase by annealing connection. Sliced PX330-HSPCAS9, PX330-ROSA26SGRNA-HSPCAS9 or PX330-GAPDHSGRNA-HSPCAS9 was obtained. The annealing connection primer w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com