Auxiliary protein for improving DNA repair efficiency and gene editing vector and application thereof
An auxiliary protein and gene editing technology, applied in the direction of stably introducing foreign DNA into chromosomes, recombinant DNA technology, genetic engineering, etc., can solve the problem of decreased virus packaging efficiency and achieve high homologous recombination efficiency and strong repair function Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] The expression plasmids of auxiliary proteins BRID, ARD and BRID-ARD were constructed respectively.
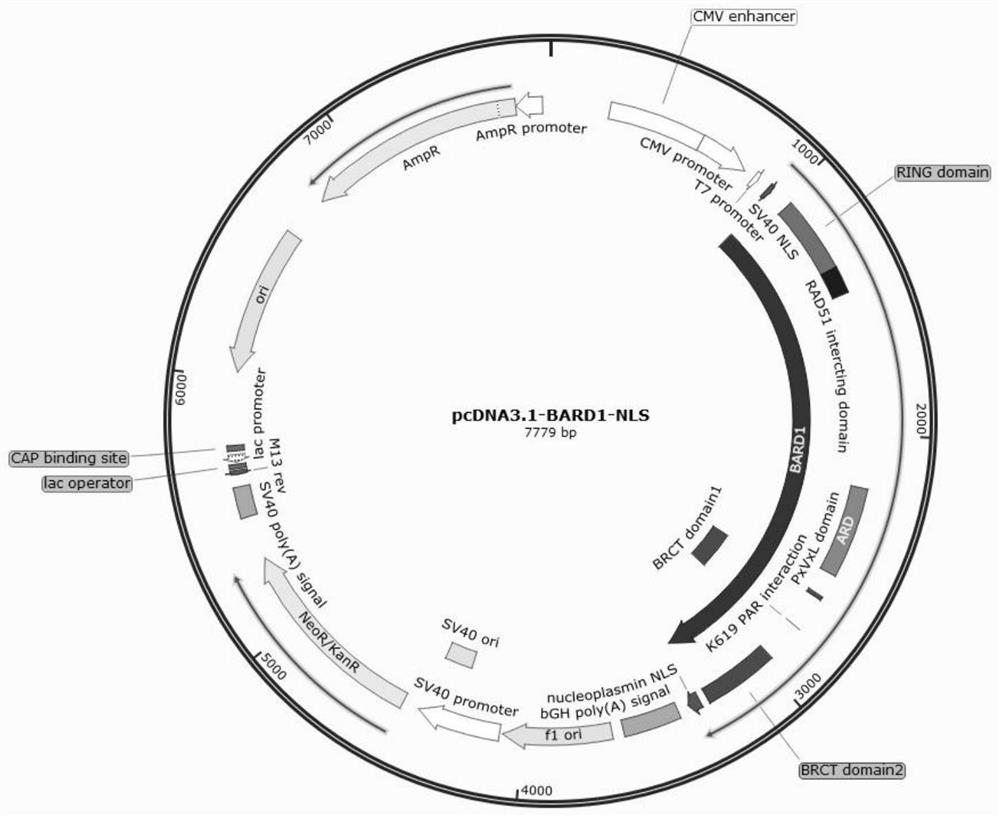
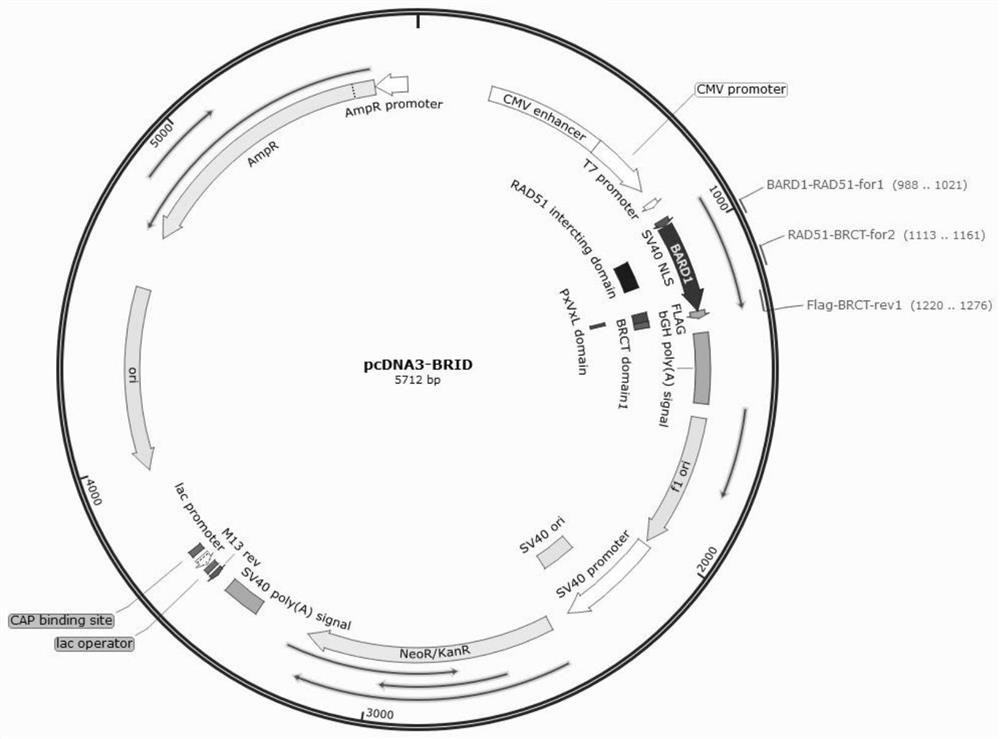
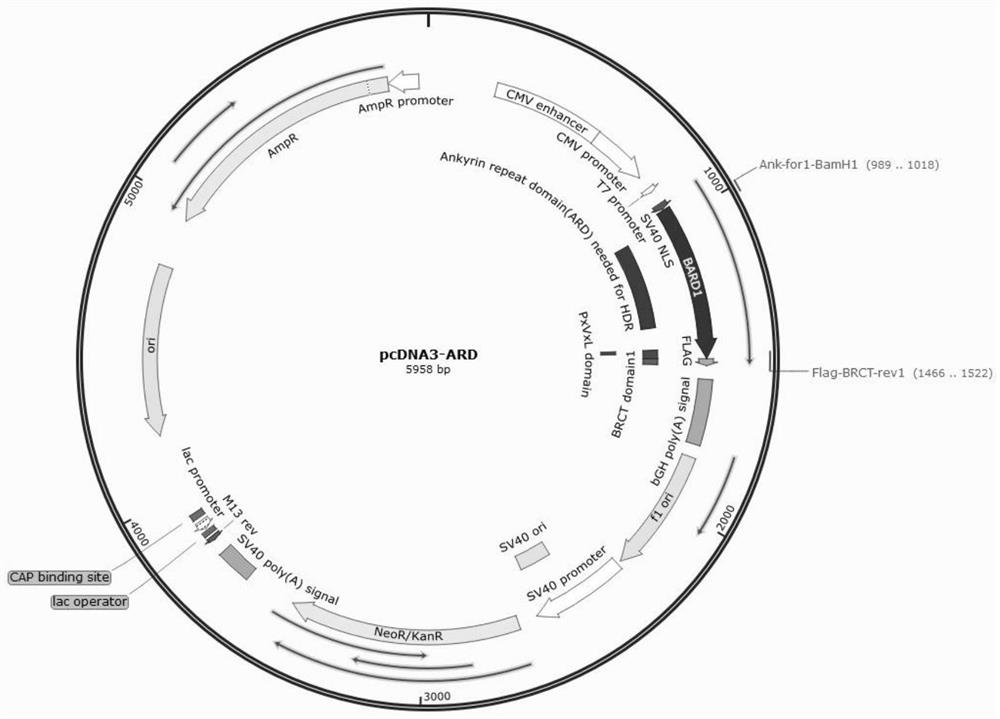
[0042] The pcDNA3-BRID, pcDNA3-ARD and pcDNA3BRID-ARD expression plasmids were constructed using the accessory protein BRID, ARD and BRID-ARD fragments as insert DNA, and the pcDNA3.1-NLS-BARD1 plasmid as the vector backbone. Wherein, the nucleotide sequences of the auxiliary proteins BRID, ARD and BRID-ARD are respectively shown in SEQ ID NO:1, SEQ ID NO:2, and SEQ ID NO:3. The plasmid map of the pcDNA3.1-NLS-BARD1 plasmid is as follows figure 1 Shown, the nucleotide sequence is shown in SEQ ID NO:7.
[0043] The construction process is as follows: take BRID, ARD and BRID-ARD auxiliary proteins respectively, use BamH1+Xho1 endonuclease (NEB company), double enzyme digestion at 37°C for 2 hours, and use a kit (Novazyme, catalog number: DC301-01) The DNA products (as insert DNA) were recovered separately; the pcDNA3.1-NLS-BARD1 plasmid was also digested with BamH1+Xho...
Embodiment 2
[0048] Under the action of CRISPR / Cas9, the fluorescent reporter plasmid pDR-GFP induces the DNA repair process of the NHEJ pathway or the HDR pathway autonomously in cells. Only by relying on the HDR repair method, the cells can normally emit green fluorescence. If the NHEJ-mediated DNA junction repair, the GFP fluorescence will be weak or no fluorescence.
[0049] In this example, the fluorescent reporter plasmid pDR-GFP (purchased from Addgene, #26475) was used as a reporter plasmid for HDR pathway-dependent DNA repair efficiency. The expression plasmids used to detect the three auxiliary proteins constructed in Example 1 have DNA repair efficiency on the HDR pathway.
[0050] Design SceGFPgRNA1 for the DNA sequence near the I-SceI restriction site on the fluorescent reporter plasmid pDR-GFP, and clone it into the Lenti-sgRNA-CRISPR-v2 vector (purchased from Addgene, #52961) through the BsmB1 site to obtain Lenti-SceGFPgRNA1 - CRISPR gene editing vector. The clone method ...
Embodiment 3
[0070] Construction of gene editing vectors including BRID, ARD or BRID-ARD
[0071] 1. Amplify the Cas9 fragment
[0072] The Cas9 fragment was amplified using the Lenti-sgRNA-CRISPR-v2 plasmid (purchased from Addgene, #52961) as a vector template.
[0073] Primer sequence:
[0074] BARD1-Cas9-5-for2: tataggcagtgggctgtcttcagaaatggacaagaagtacagcatcggc (SEQ ID NO: 10);
[0075] BARD1-Cas9-5-rev2: taacgcgttcacttatcgtcatcgtctttgtaatcgtcgcctcccagctgagacag (SEQ ID NO: 11).
[0076] A high-fidelity PCR kit (Novazyme, Cat. No. P505-d1) was used to amplify. The amplification system is shown in Table 5, and the amplification procedure is shown in Table 6:
[0077] Table 5 Amplification system for amplifying Cas9 fragments
[0078] Composition Dosage 2×Phanta Max Buffer 25μl dNTP (10mMeach) 1μl Primer F (primer Cas9-5-for2) 2μl Primer R (Primer Cas9-5-rev2) 2μl DNA template (plasmid, Lenti-sgRNA-CRISPR-v2) 1ng Phanta Max Super-Fideli...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com