Method for preparing DNA vaccine with herpes simplex virus type I UL5 gene deletion
A herpes simplex virus and DNA vaccine technology, applied in the field of biomedicine, can solve the problems of complex preparation methods of attenuated live vaccines, difficulties in PCR amplification, and low probability of success, etc., and achieve virus titer and infectivity weakened, high Immunogenicity, efficiency-enhancing effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] The construction of the eukaryotic cell line of embodiment 1UL5 gene expression vector
[0041] Using the BAC-17.37 shuttle plasmid as a template, design primers (SEQIDNO.5, SEQIDNO.6) to amplify the UL5 gene (2696bp), and subclone the gene fragment into pCDNA3.1 after digestion with BamHI and BstXI restriction endonucleases (+) Plasmid (purchased from Youbao Biological Company, Cat. No. VT1001) to obtain pCDNA3.1-UL5 plasmid;
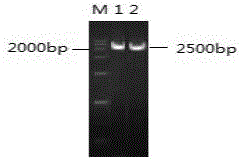
[0042] The Cre gene fragment was amplified by reverse transcription of the P1 phage RNA, the pCDNA3.1-UL5 plasmid and the Cre gene fragment were digested with BamHI and ligated with T4 ligase, and a single clone was selected for Cre gene and UL5 gene PCR identification and sequencing, the results are as follows figure 1 and figure 2 As shown, the eukaryotic expression pCDNA3.1-UL5-CRE plasmid was obtained;
[0043] The pCDNA3.1-UL5-CRE plasmid was transfected into Vero cells (purchased from ATCC, USA) using lipo2000 liposomes (purchased from...
Embodiment 2
[0045] The homologous recombination knockout of embodiment 2HSV-1UL5 gene
[0046] 2.1 Construction of pREDI recombinant plasmid
[0047] Design the PCR amplification primers of the promoter PrhaB, and obtain the PrhaB fragment (2.0kb) through PCR amplification; design the PCR amplification primers of I-SceI, and obtain the I-SecI fragment (0.7kb) through PCR amplification; The I-SecI fragment is connected by recombinant PCR to obtain the PrhaB-I-SecI fragment; the PrhaB-I-SecI fragment is connected to the pKD46λ-redrecombinase vector to obtain the pREDI recombinant plasmid. The pREDI recombinant plasmid has λ-redrecombinase induced by L-arabinose and I-SecI endonuclease gene induced by rhamnose.
[0048] 2.2 Construction of recombinant knockout system
[0049] The constructed pREDI recombinant plasmid was transformed into a host bacterium containing the BAC-HSV-1 vector, and the host bacterium was EPI300E.Coli to obtain a recombinant knockout system for homologous recombina...
Embodiment 3B
[0056] The excision of embodiment 3BAC carrier and the acquisition of HSV-1UL5 gene-deficient strain
[0057] The recombinant strain without UL5 gene and foreign gene was expanded and cultivated, and the BAC mass extraction kit was used for plasmid extraction to obtain the BAC-17.37ΔUL5 plasmid, which was electrotransformed into Vero-pCDNA3.1 constructed in Example 1 - UL5-Cre cells were cultured in DMEM medium containing 2% by volume of fetal bovine serum and 1ug / ml of G418. 24 hours before the occurrence of cell lesions, the cell culture medium was removed, and then cultured with 1% agar Sugar and 2% FBS were covered with DMEM medium. When visible pathological effects were formed, phosphate buffer containing 400ug / ml X-gal (purchased from BioSynth AG, Switzerland) was added, and then the white spots were selected by the blue-white spot test, repeatedly frozen and thawed, and centrifuged The supernatant was used to obtain the HSV-1 UL5 gene-deleted DNA vaccine.
[0058] Infe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com