Method and system for predicting interaction between miRNA and gene based on multi-relational graph convolutional network
A convolutional network and relational graph technology, applied in deep learning in the field of bioinformatics, can solve problems such as relying on manual extraction, achieve the effect of improving prediction accuracy and enriching prediction methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
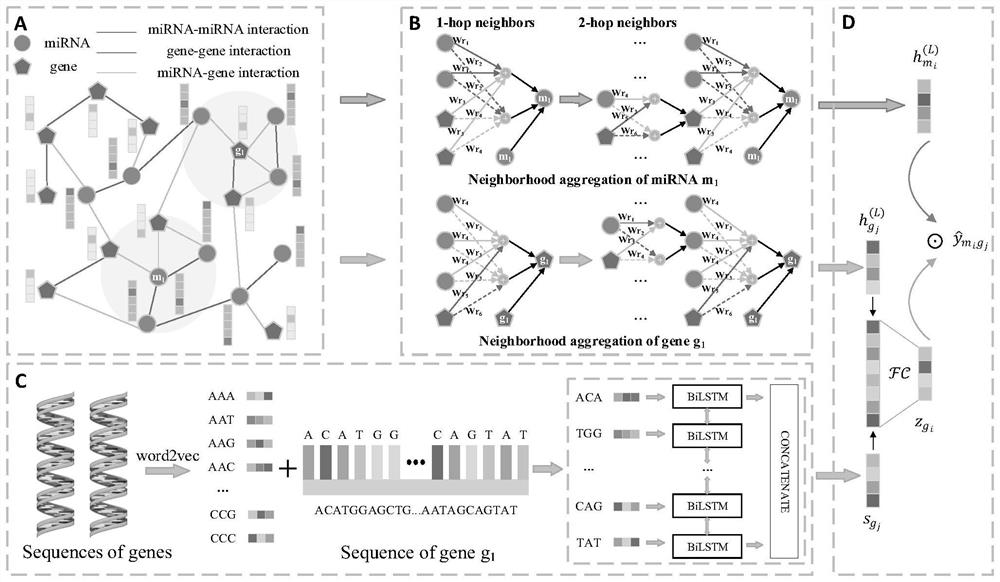
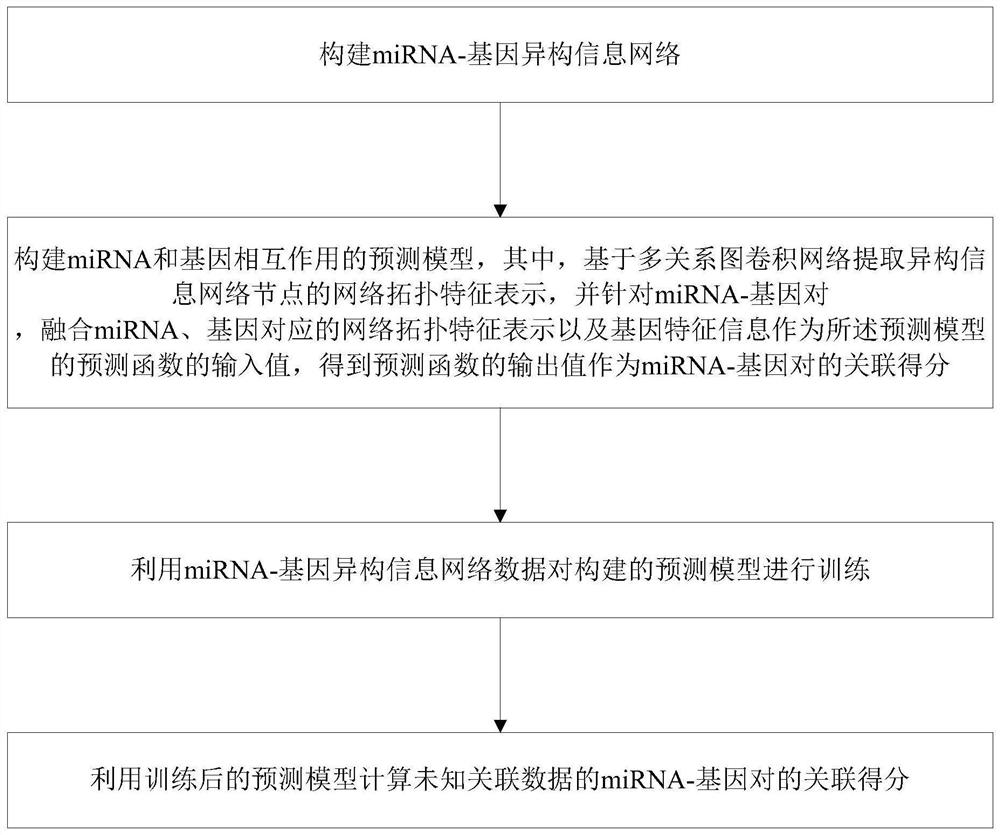
[0057] Such as figure 1 As shown, the prediction method of miRNA and gene interaction based on a multi-relational graph convolutional network provided in this embodiment includes the following steps:
[0058] Step S1: Construction of miRNA-gene heterogeneous information network. Among them, miRNA-gene heterogeneous information network is obtained by integrating miRNA similarity network, gene similarity network and known miRNA-gene bipartite network.
[0059] In this embodiment, the miRNA sequence is obtained from the miRBase database, and the sequence similarity scores between all miRNA-miRNA pairs are calculated by the overall sequence comparison algorithm Needleman-Wunsch, and the 10 most related neighbor nodes are reserved for each miRNA, namely The 10 miRNA-miRNA relationship data with the highest similarity score were retained, and then the miRNA similarity network was constructed. It should be understood that the present invention is not limited to retaining 10 neighbo...
Embodiment 2
[0104] Embodiment 2: This embodiment provides a system based on the prediction method of the above-mentioned miRNA and gene interaction, which includes:
[0105] The miRNA-gene heterogeneous information network building block is used to construct the miRNA-gene heterogeneous information network, and the miRNA-gene heterogeneous information network includes relationship data between miRNA-miRNA and association data of known miRNA-gene pairs , gene-gene relationship data;
[0106] The prediction model building block is used to construct the prediction model of miRNA and gene interaction, in which, based on the multi-relational graph convolutional network, the network topology feature representation of the heterogeneous information network nodes is extracted, and for the miRNA-gene pair, the miRNA and gene correspondence are fused The network topology feature representation and gene feature information are used as the input value of the prediction function of the prediction model...
Embodiment 3
[0112] This embodiment provides an electronic terminal, which includes:
[0113] one or more processors;
[0114] memory storing one or more computer programs;
[0115] The processor invokes the computer program to:
[0116] Steps in a method for predicting miRNA and gene interactions based on multi-relational graph convolutional networks. Implementation:
[0117] Step S1: Construction of miRNA-gene heterogeneous information network.
[0118] Step S2: Construct a predictive model of miRNA-gene interaction.
[0119] Step S3: Using the miRNA-gene heterogeneous information network data in step S1 to train the prediction model constructed in step S2.
[0120] Step S4: Using the trained prediction model to calculate the association score of miRNA-gene pairs with unknown association data.
[0121] For the specific implementation process of each step, please refer to the description of the foregoing method.
[0122] It should be understood that in the embodiment of the present...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com