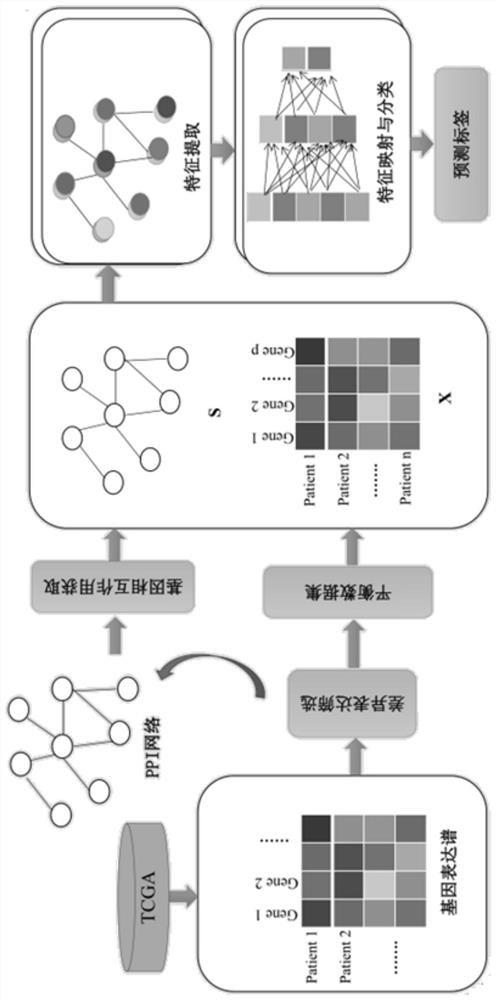
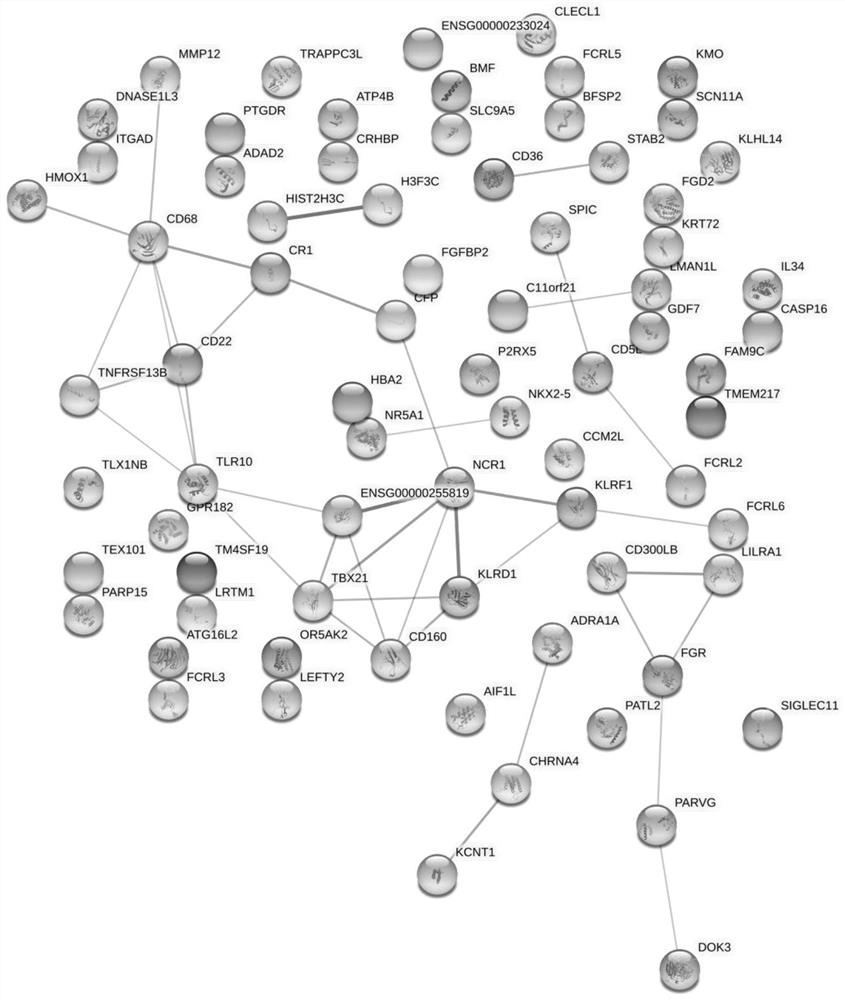
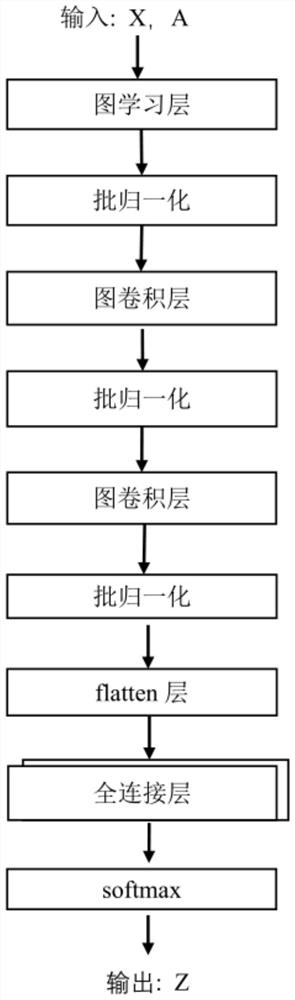
Distance metastasis identification method based on gene interaction mode optimization graph representation
An identification method, a technology of distant transfer, applied in the field of biological information, can solve problems such as incompatibility, and achieve the effect of accurately predicting performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0123] For the CESC sample data set, follow the method of step 5 above to input the training set CESC_Xtrian of each fold described in step 3-1 into the glmGCN model based on the optimization graph representation of the gene interaction mode described in step 4, by minimizing step 4 The loss function described in -4 is optimized to obtain the network weight W_CESC. And use step 5-2, step 5-3 and step 5-4 to obtain the average value of the ten-fold cross-validation results of three repeated experiments as the final result.
[0124] For the STAD sample data set, according to the method of step 5 above, the training set STAD_Xtrian of each fold described in step 3-2 is input into the glmGCN model based on the optimization graph representation of the gene interaction mode described in step 4, and by minimizing step 4 The loss function described in -4 is optimized to obtain the network weight W_STAD. And use step 5-2, step 5-3 and step 5-4 to obtain the average value of the ten-fo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com