Biomarker for breast cancer typing and application thereof
A biomarker and breast cancer technology, applied in the field of medical detection, can solve problems such as poor differentiation and pathological judgment errors, and achieve the effect of convenient and precise treatment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Based on the TCGA public database, a preliminary screening of variant gene markers for pathological subtypes of breast cancer was carried out.
[0060] The screening method is as follows.
[0061] 1. Obtain whole exome sequencing data of tumor tissue of breast cancer patients from TCGA database.
[0062] In this example, a total of 705 cases of breast cancer patients (including 490 cases of invasive ductal carcinoma and 215 cases of invasive lobular carcinoma) were downloaded from whole exome sequencing data, and seven different software were used: Samtools, SomaticSniper, Strelka and VarScan to detect points respectively Mutations; InDels were detected by VarScan, Pindel, GATK and Strelka, respectively.
[0063] 2. According to the difference analysis between the invasive ductal carcinoma group and the lobular carcinoma group.
[0064] The chi-square test was used for statistical analysis, and the variant genes with p≤0.05 and the genes investigated in the literature...
Embodiment 2
[0076] The latent markers obtained in Example 1 were used to train the model.
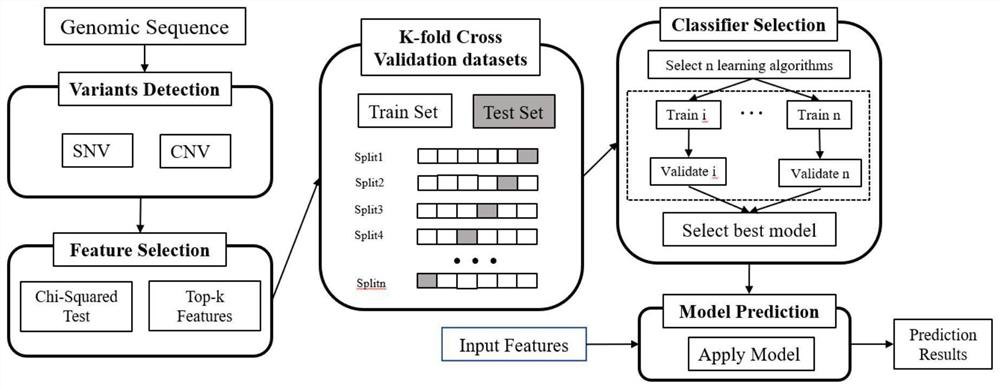
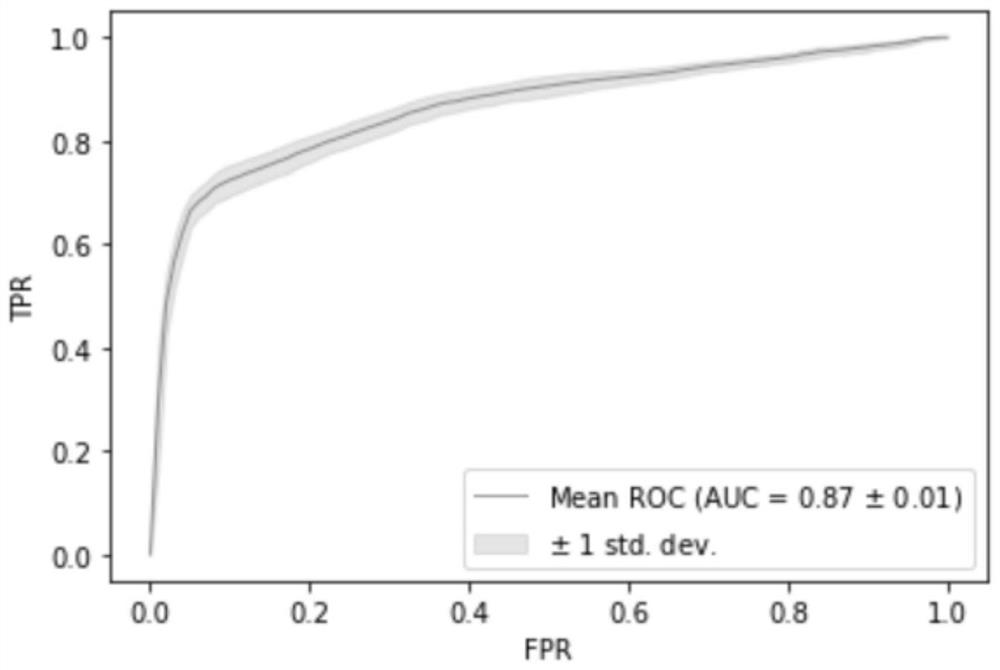
[0077]1. Using the information of all target markers obtained in Example 1, the TCGA and breast_msk_2018 data sets were combined to train the model, and one sample lacking CNV data was removed, that is, 1962 cases of ductal carcinoma and 603 cases of lobular carcinoma patient tissue samples for detection and judgment , using the random forest model for modeling analysis, the modeling process is as follows figure 1 As shown, according to the 7:3 segmentation, 20 repetitions are performed, and the model AUC is as high as 0.8685, such as figure 2 shown.
[0078] 2. Optimizing 111 markers: By using the random forest model for modeling analysis, 1962 cases of ductal carcinoma and 603 cases of lobular carcinoma were detected and judged, and 20 repetitions were performed according to the division of 7:3. The feature importance of the model in step 1, select the optimal combination of the top 20 MARKER,...
Embodiment 3
[0082] The biomarkers in Example 1 and the model in Example 2 were verified.
[0083] The verification process is as follows.
[0084] 1. Obtaining tissue samples: 897 cases of breast cancer (660 cases of invasive ductal carcinoma and 237 cases of invasive lobular carcinoma) were collected from Jinan University and related FFPE section samples were identified by relevant experts.
[0085] 2. Sample sequencing analysis:
[0086] FFPE tissue samples were analyzed by whole genome sequencing by a third party (Clearcode Biotechnology).
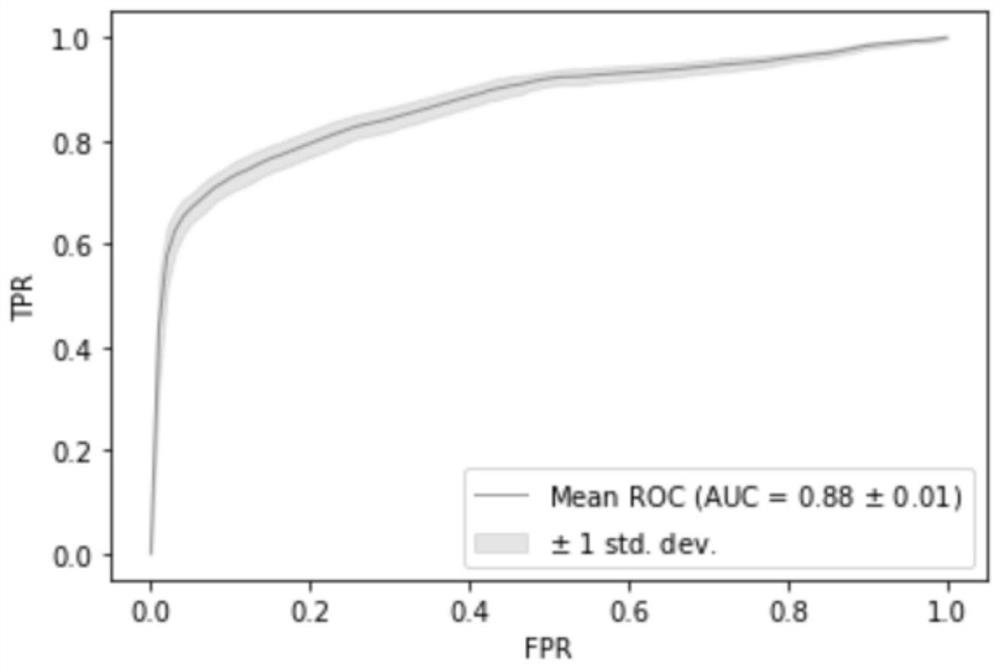
[0087] 3. Use the above 111 marker information to detect and judge the independent verification set, that is, patient tissue samples. According to the prediction test of the 20 marker models obtained in Example 2, the AUC in Example 3 can reach 0.9048, as Figure 6 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com