Disease drug prediction method based on heterogeneous network embedding model
A heterogeneous network and prediction method technology, applied in drug reference, genomics, bioinformatics, etc., to achieve the effect of reducing research and development costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0064] The specific implementation of the present invention will be described in detail below in conjunction with the technical scheme and accompanying drawings.
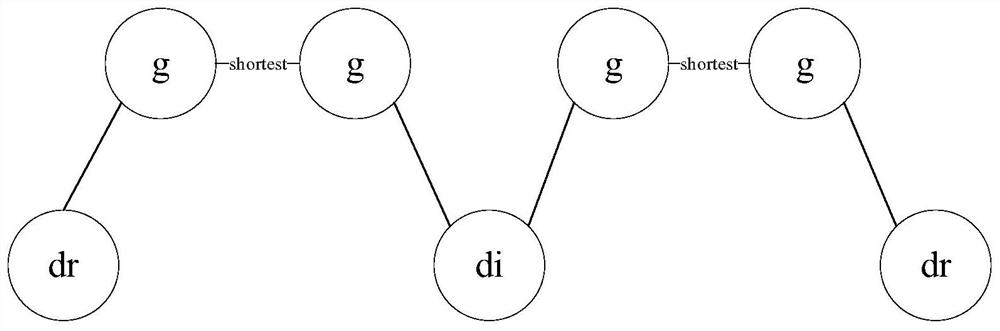
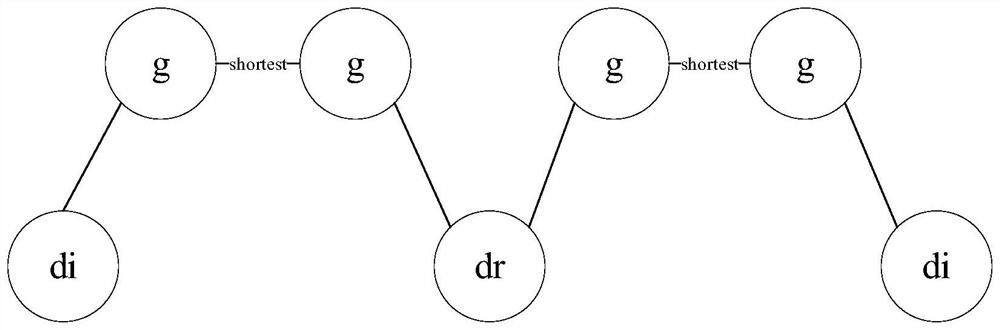
[0065] There are 141,296 protein-protein interaction data consisting of 13,460 proteins, 299 diseases and the corresponding OMIM and GWAS gene data involved, 238 drugs from DrugBank and corresponding target data, and 403 diseases Relationship data with drugs.
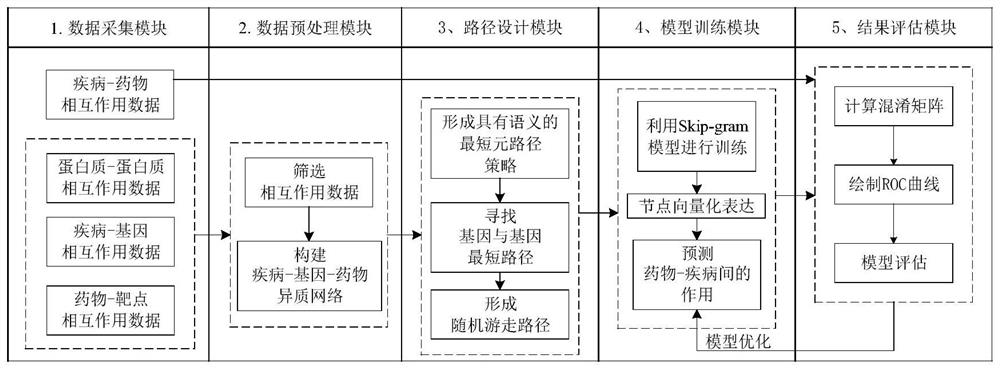
[0066] Such as figure 1 As shown, a disease drug prediction method based on heterogeneous network embedding includes data acquisition module, data preprocessing module, path design module, model training module, and result evaluation module, as follows:
[0067] (1) The data acquisition module includes:
[0068] (1-1) Interaction data between diseases and drugs: the collected drug data has a corresponding therapeutic effect on the diseases involved, and this data will be used as a test set to verify the interaction between the predicted diseases and drugs ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com