SNP (Single Nucleotide Polymorphism) marker for cloud black goats and application of SNP marker in identification of varieties of cloud black goats
A black goat and identification cloud technology, which is applied in the direction of recombinant DNA technology, microbial measurement/inspection, DNA/RNA fragments, etc., can solve the problems of time-consuming, and the accuracy of results needs to be improved.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Example 1 First screening of cloud-shang black goat differential SNP sites
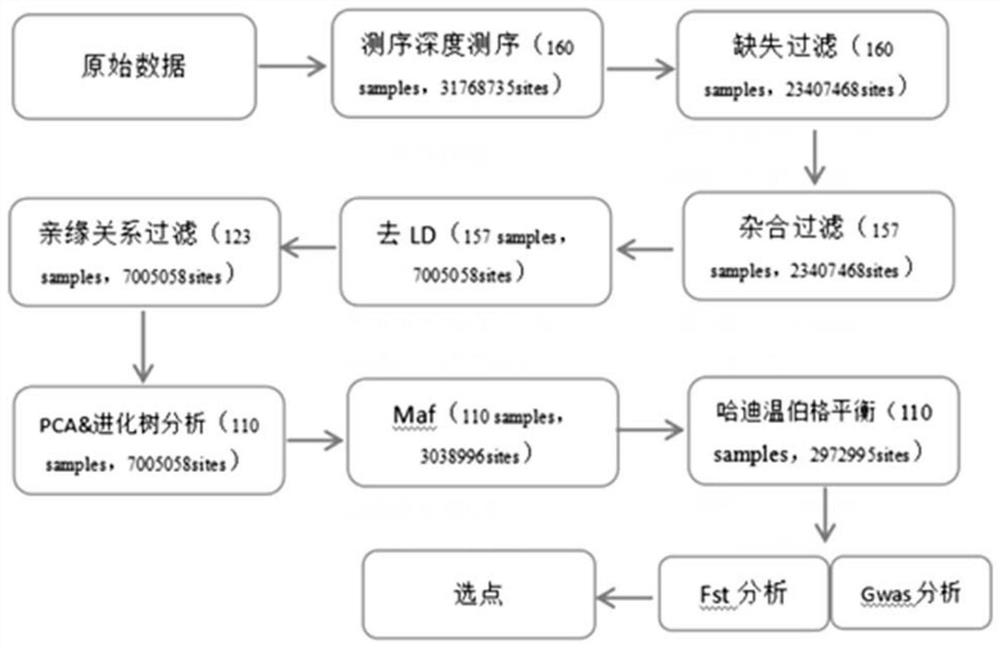
[0057] For 4 breeds (Yunshang black goat, hybrid sheep F2 generation, Yunling black goat and Chuanzhong black goat), a total of 160 sample data, including 31768735 loci. In order to obtain high-quality differential loci between varieties, the samples and loci are first filtered, and then the differential loci are screened on this basis. The specific flowchart is as follows figure 1 .
[0058] GWAS analysis and Fst analysis were performed on the 110 samples retained after filtering and the remaining loci after quality control, so as to screen the differential loci among varieties. In this project, there are 4 breeds, so in gwas analysis and Fst analysis, we divide the samples into 2 groups (for example, YSH.vs.Other, YSH: black goat on the cloud), and obtain significant difference sites respectively. Based on the results of GWAS and Fst, the TOP 23 SNP sites with significant differences betwee...
Embodiment 2
[0091] Example 2 Second screening of cloud black goat differential SNP sites
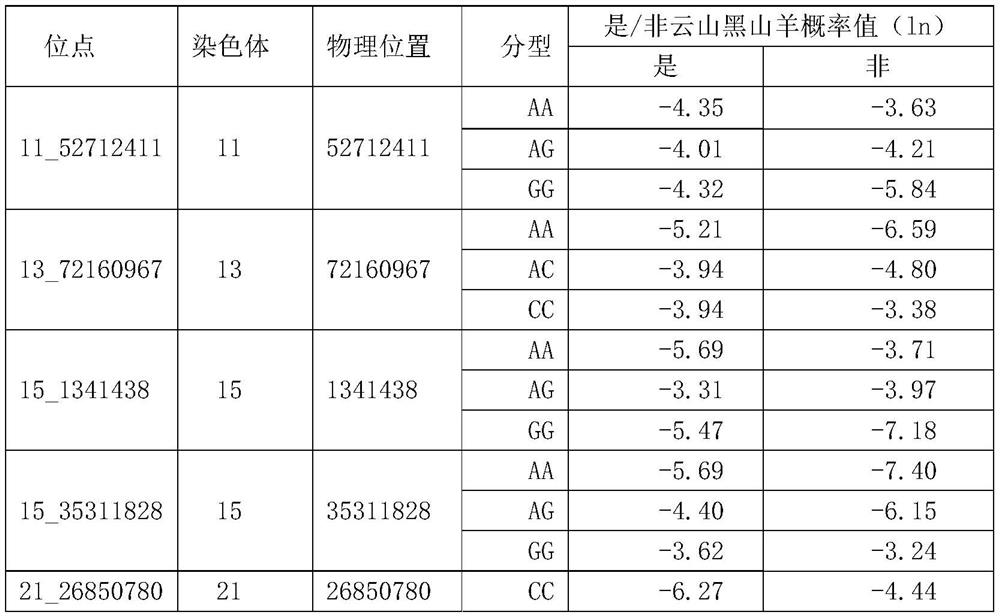
[0092] Considering the identification accuracy rate and the distribution of loci in the whole genome, the second selection is carried out according to the analysis results of the resequencing data. The screening principle is to select loci with large differences between varieties, and try to ensure that the loci are evenly distributed in the whole genome. Distribution, the newly selected 26 loci are distributed on 12 chromosomes such as 3, 4, 5, etc. The detailed information of the loci is shown in the table below.
[0093] Table 3 26 SNP sites for the second screening
[0094]
[0095]
[0096] Library building and blind testing:
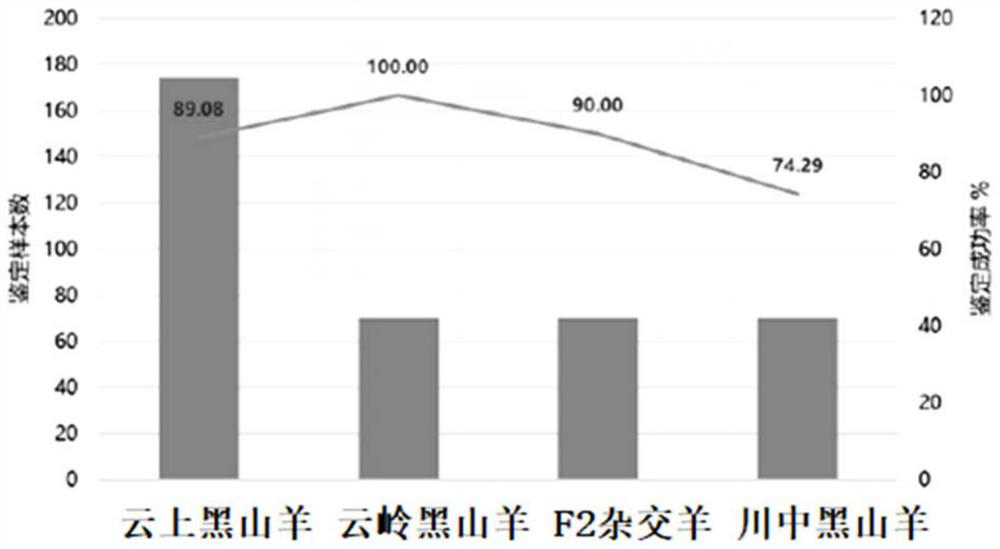
[0097] The genotyping of 26 loci of 384 samples of 4 varieties was detected by mass spectrometry, and the blindly tested samples were identified after the completion of the library construction. For the specific statistical results, see figure 2 .
[0098] Depe...
Embodiment 3
[0100] Example 3 Construction of a reference group data model for identifying cloud black goats and non-cloud black goats
[0101] Collected 1013 cloud black goats (Shuangbai County Yunnan Xianghong Agriculture and Animal Husbandry Co., Ltd., self-employed households in Shiyang Town, Dayao County, etc.) and 1291 non-cloud black goats (Shuangbai County Ainishan Haizidi Village, Dayao County, etc.) County Sanchahe Township, Sichuan Ziyang City Lezhi County Chuanzong Sheep Industry Company, etc.) blood or ear tissue genomic DNA, and then Sequenom test. Among them, the specific operation steps of Sequenom detection are as follows:
[0102] 1. Primer Design
[0103] Use Genotyping Tools and MassARRAY Assay Design software of Sequenom company to design PCR amplification primers and single-base extension primers of the SNP site to be tested, and hand them over to the bio-company for synthesis. The primer information for each SNP is shown in the sequence 1-79 in Table 4:
[0104] T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com