Library building connector and method suitable for simplified genome sequencing of DNBSEQ technology
A technology of genome sequencing and technology, applied in the field of bioinformatics, can solve problems such as the complexity of database construction methods, and achieve the effect of simplifying genome sequencing and high sequencing quality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
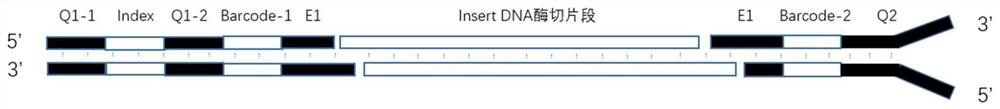
[0034] This example designs a linker for library construction, the linker is composed of one or several pairs of DNA double-stranded fragments, and each pair of linkers is divided into an upstream linker sequence (F) and a downstream linker sequence (R).
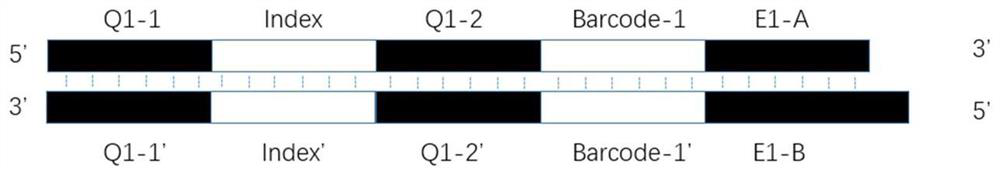
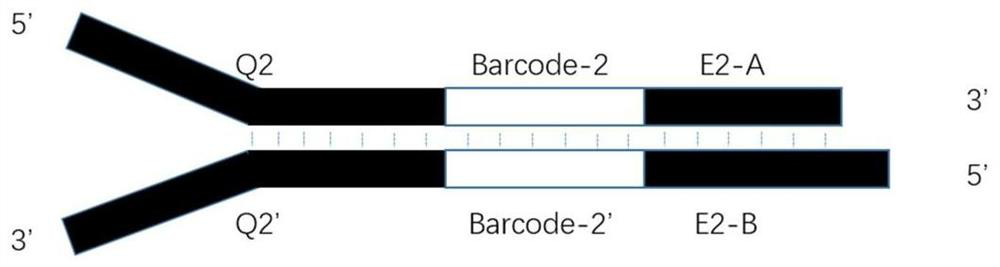
[0035] like figure 1 As shown, the upstream linker F contains the following structure:
[0036] (1) An enzyme cutting end portion (E1) used for complementary pairing with a sticky end produced by a restriction endonuclease, and the E1 sequence can be replaced according to the selected restriction endonuclease, such as a linker sequence. E1 is a sticky end, which contains two DNA single strands, forward strand E1-A and reverse complementary strand E1-B. Among them, E1-A from 5' to 3' direction is the double-stranded partial sequence in the sticky end left after the corresponding restriction endonuclease digests DNA; E1-B from 3' to 5' direction is the restriction endonuclease The reverse strand sequence left after enzymatic...
Embodiment 2
[0052] This example uses the sequencing adapter shown in Example 1 to design a simplified genome sequencing method suitable for DNBSEQ technology. The specific process is as follows:
[0053] 1. Adapt to different restriction enzymes and Barcode / Index
[0054] According to the linker structure and general formula designed in this invention, the linker structure can be designed for the following sequencing requirements:
[0055] (1) When genomic DNA was digested with EcoRI-MspI double enzyme, library construction was performed for 3 individuals. 3 kinds of Index, 3 kinds of Barcode-1, 3 kinds of Barcode-2 are needed in order to meet the sequencing requirements, and the E1 and E2 fragments are respectively replaced by the enzyme cut sticky ends of EcoRI and MspI, specifically as shown in Table 2 (SEQ ID NO.7- 18):
[0056] Table 1: When using EcoRI-MspI double restriction enzyme to digest genomic DNA, 3 individual Barcodes, Index and restriction sites
[0057]
[0058] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com