Method of recombination and agents useful for same
A homologous recombination and inhibitor technology, applied in the field of recombining circular nucleic acid sequences and linear nucleic acid sequences and modifying bacterial artificial chromosomes, can solve problems such as the degree of homologous recombination that cannot be well controlled
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0117] Example 1 Inducible recE pathway for homologous recombination of PCR fragments to circular substrates in E. coli DH10B cells
[0118] The recE pathway for homologous recombination between linear and circular substrates is active in sbcA recBC strains JC8679 and JC9604 42-45 . The pathway is independent of recA 46 , because the recT gene seems to fulfill the role of recA in pairing with homologous sequences 47 . In these cells, kanamycin resistance (Km r ) PCR fragment of gene (PCRkan50) was precisely homologously recombined into chloramphenicol resistance (Cm r ) in the backbone of the pEBAC140 vector, resulting in pEBAC140::kan50. However, it was not possible to transfer the 200 kbp EBAC / 148-globin clone in DH10B cells to JC9604 electrocompetent cells for subsequent modification, presumably due to host-restricted differences between the two strains. Since BAC / PAC are normally maintained in DH10B cells, any modification can be performed directly in DH10B cells har...
Embodiment 2
[0121] Example 2 Homologous recombination of PCRkan50 and PCRkan140 into the vector backbone of the 200 kb β-globin clone pEBAC / 148 in DH10B cells
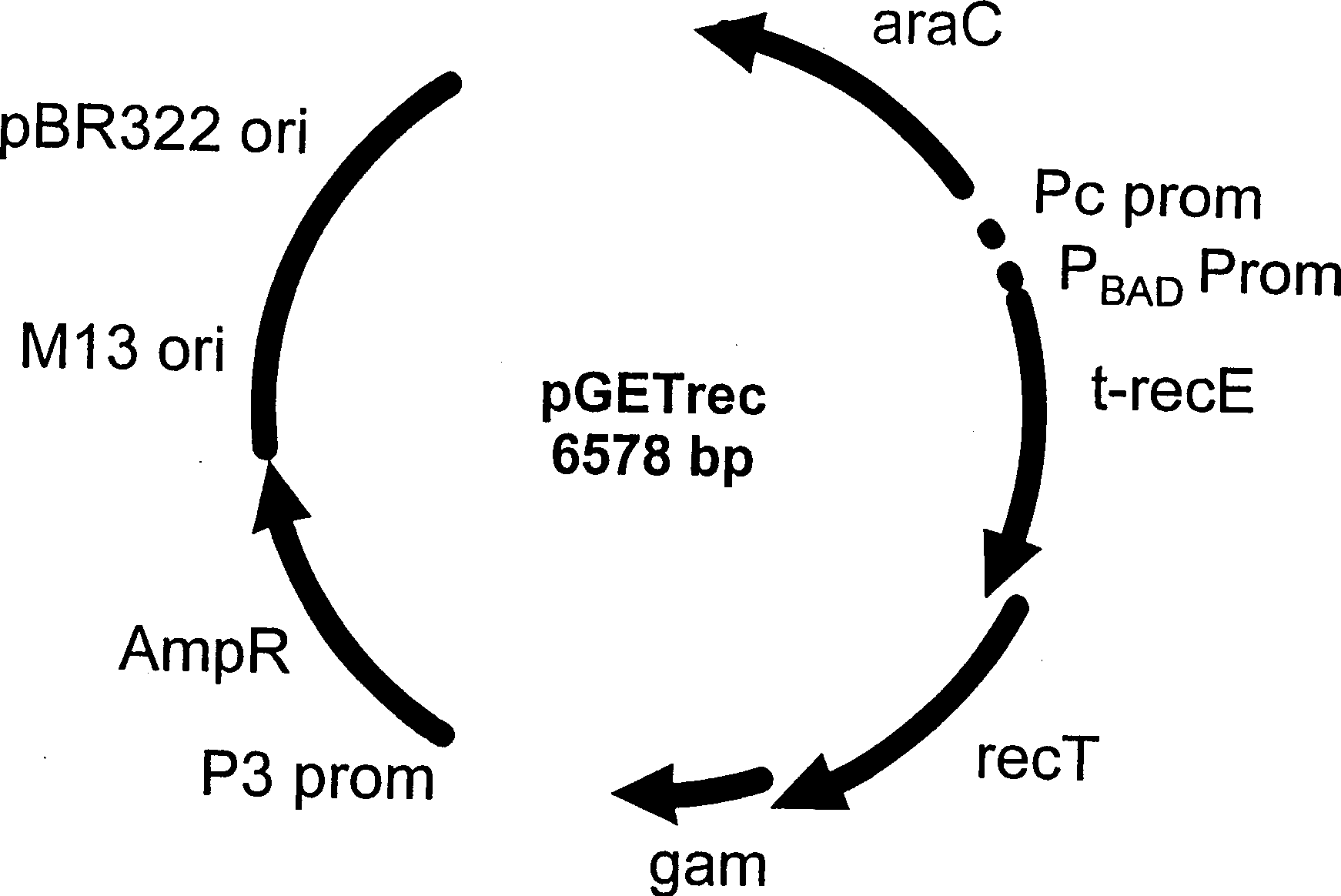
[0122] We attempted to modify the 200 kb β-globin clone pEBAC / 148 using PCRkan140 to see if the same method could be used to modify a large, single-copy BAC clone. pGETrec was electroporated into DH10B (pEBAC / 148) cells. Electrocompetent cells were prepared using clone DH10B (pGETrec, pEBAC / 148) carrying both plasmids. Cells were induced with L-arabinose for 40 minutes, and then collected to prepare electrocompetent cells. Electroporation of PCRkan140 into these cells yielded over 1000 Cm per electroporation r Km rRecombinants (Table 1, Experiments 3, 4). When the cells were induced with L-arabinose at the beginning of the culture before the preparation of electrocompetent cells, no Cm r Km r Recombinant. Under L-arabinose induction, prolonged overexpression of recE, recT and gam genes was toxic to DH10B cells. Cells harbo...
Embodiment 3
[0129] Example 3 Construction of pEBAC / 148
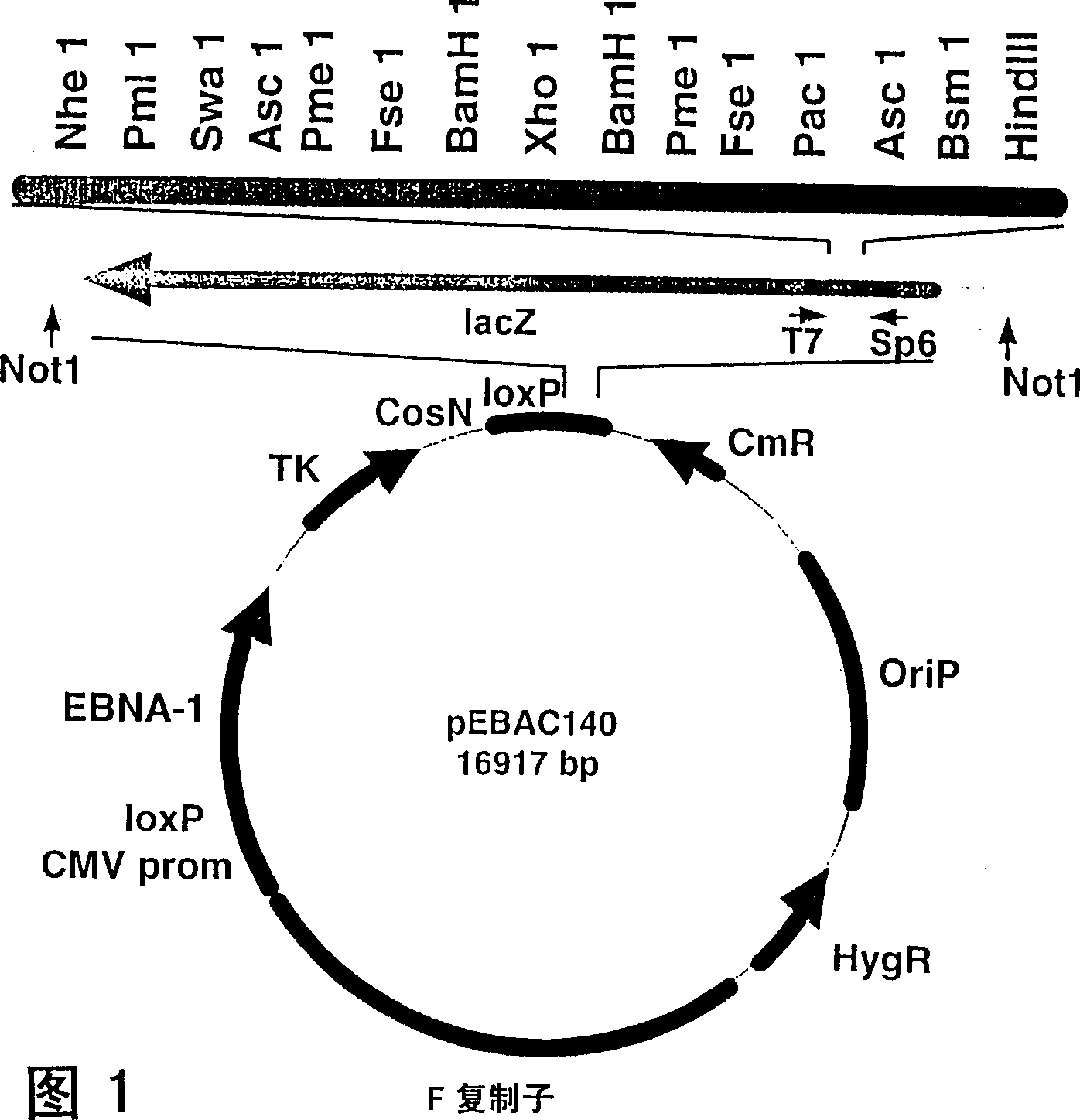
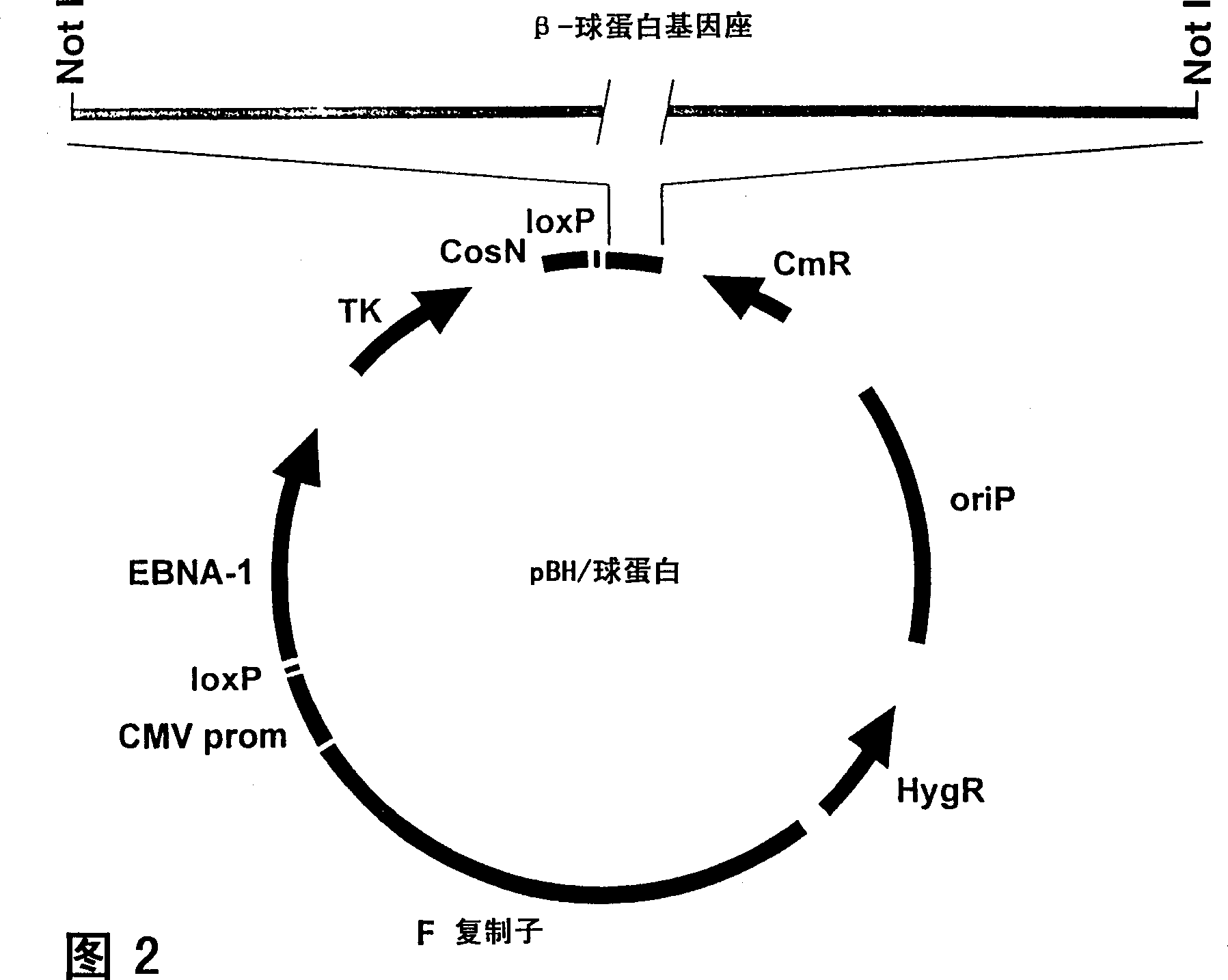
[0130] The second generation pEBAC140 BAC / PAC cloning vector (Figure 1) pools the first generation PACs 24 and BAC 23 Multiple features of cloning systems, and HAEC systems 48 OriP and EBNA-1 from Epstein-Barr virus. It also contains several other features, including polyclonal regions of rare cuts. pEBAC / 148 (Fig. 2) was made from pEBAC140 by reverse ligation of a 185 kb genomic fragment carrying the entire β-globin locus using a method based on the PAC cloning protocol. Briefly, the RPCI 1 human total genome PAC library was screened by PCR primers derived from the 5'- and 3'-ends of the β-globin locus 49 . A PAC clone (PAC148 / β-globin) carrying a 185 kb genomic insert was identified and confirmed to contain the complete β-globin locus (about 73 kb), as well as additional sequences at the 5' and 3' ends. The 185 kb genomic insert was separated as a single fragment by NotI digestion, pulsed field gel electrophoresis ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com