Structure of G-protein (RGS4) and methods of identifying agonists and antagonists using same
An antagonist and agonist technology, applied in chemical instruments and methods, testing pharmaceutical preparations, animal/human peptides, etc., can solve problems such as reduced motility of Giα1 transition region
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] G protein, heterotrimeric guanine nucleotide-binding protein; RGS4, regulator of G protein signaling; G iα1 , the Gα subunit of the heterotrimeric G protein, G iα1 -AIF 4 - , with Mg 2+ , GDP and AlF 4- Complex Gα subunit of heterotrimeric G protein stabilized in GTP hydrolysis transition state, DTT, DL-1,4-dithiothreitol; GTP, guanosine triphosphate; GDP, guanosine diphosphate; NMR, nuclear magnetic resonance; 2D, two-dimensional; 3D, three-dimensional; HSQC, heteronuclear single-level quantum coherence spectroscopy; HMQC, heteronuclear multilevel quantum coherence spectroscopy; TPPI, time-proportional phase increment; Nuclear Overhauser effect; NOESY, nuclear Overhauser enhanced spectroscopy; COZY, correlation spectroscopy; HNHA, amide proton to nitrogen to CαH proton correlation (amide proton to nitrogen to CαH proton HNHB, amide proton to nitrogen to CβH proton correlation (amide proton to nitrogen to CβHproton correlation); CT-HCACO, constant time CαH proton t...
Embodiment 2
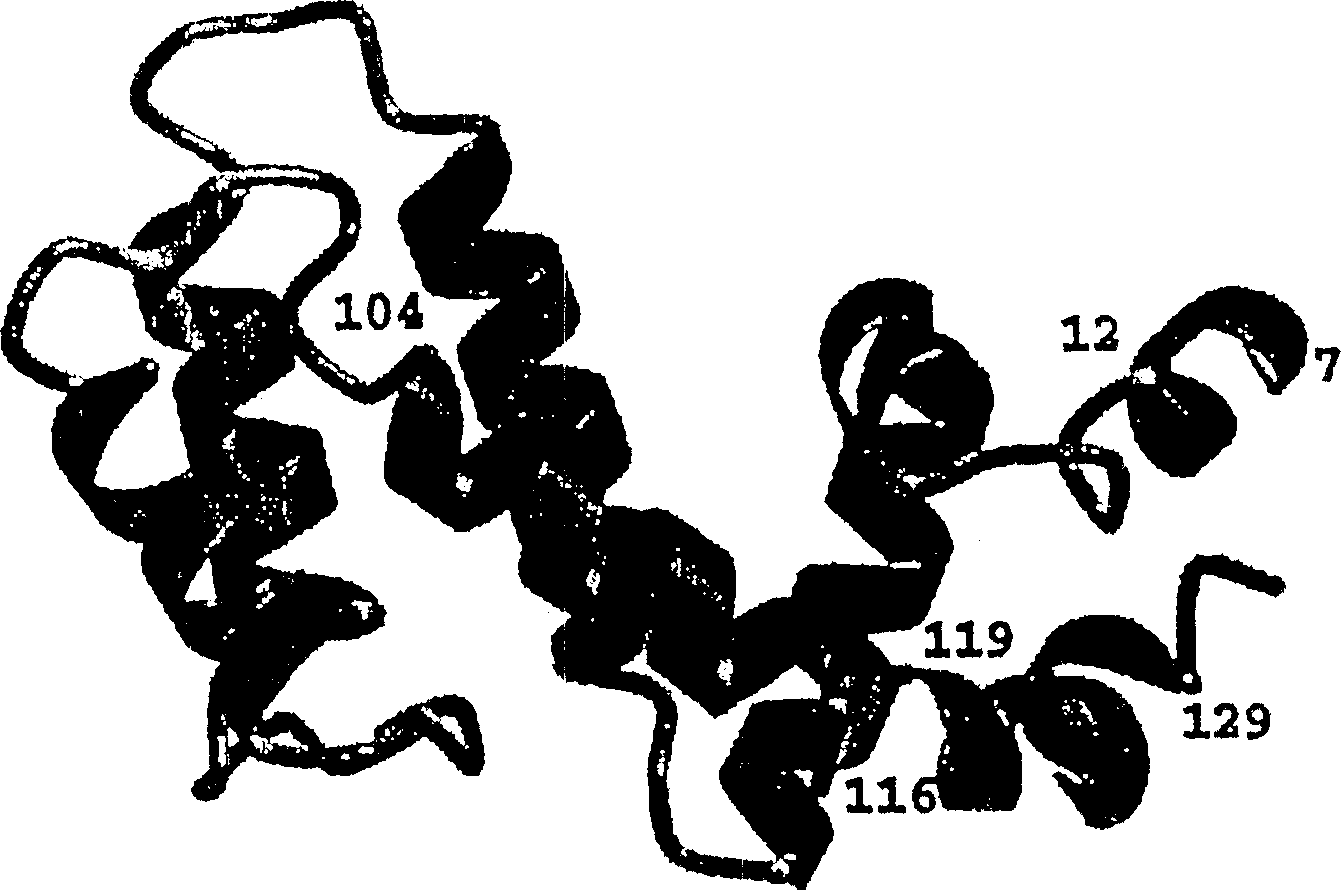
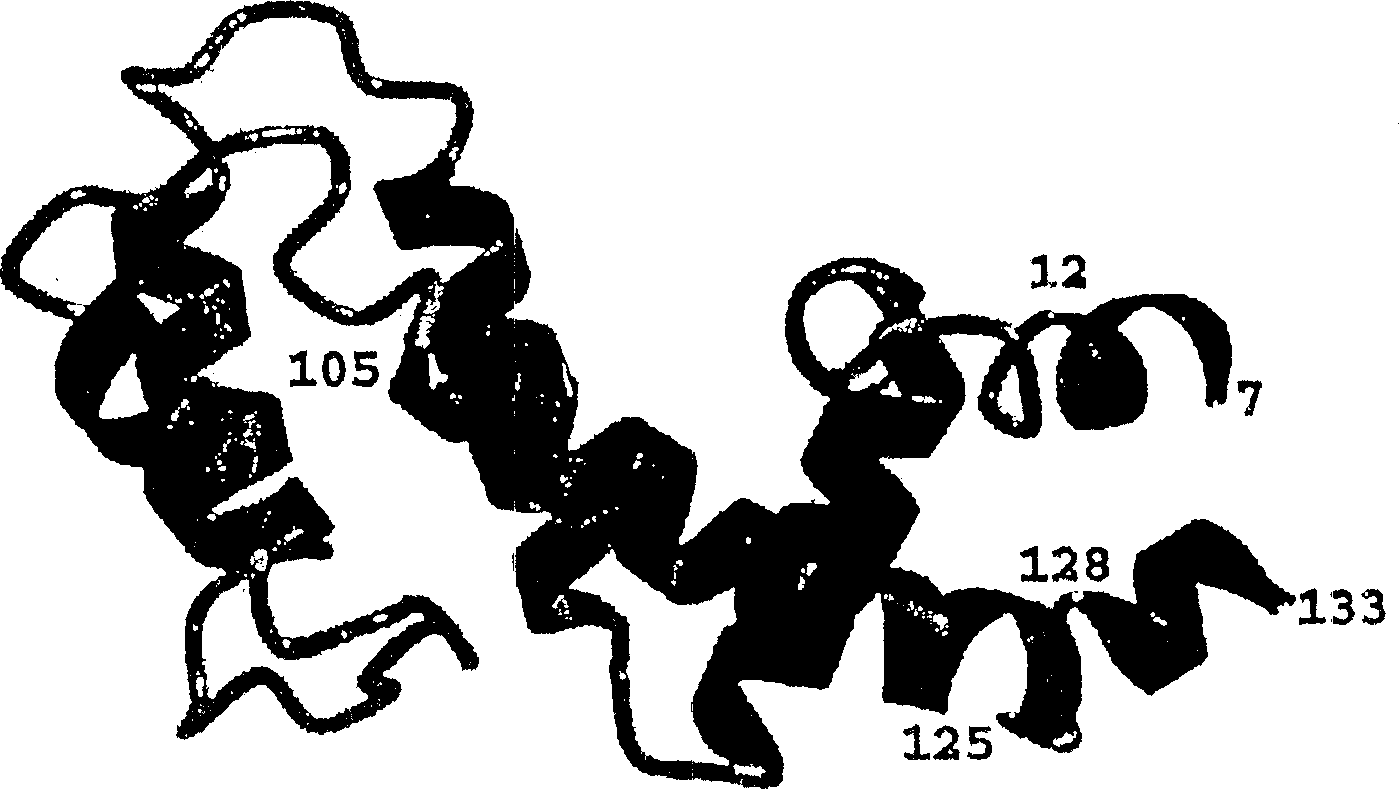
[0075] The secondary structure of the RGS4-core (outlined in Figure 1) was obtained from 15 N-modified NOESY-HMQC and 13 C - Characteristic NOE data of NH, Hα and Hβ protons of the corrected NOESY-HMQC spectrum, obtained from HNHA 3J HNα coupling constants, slowly exchanging NH protons and 13 Cα and 13 Cβ secondary chemical shifts (for review see: (56) and (78)). RGS4-core solution NMR has been determined to consist of seven helical regions corresponding to the following residues: residues 7-12 (α1); 17-36 (α2); 40-53 (α3); 61-71 (α4); 86-95 (α5); 105-125 (α6) and 128-132 (α7). The RGS4-core overall fold essentially consists of two 4-helical bundles, both of which have a long helical region α6 portion. A clear difference between the RGS4 core secondary structure in solution and the X-ray structure of the RGS4-Giα1 complex was unexpectedly observed at the C-terminus. The X-ray structure shows that residues 104-116 and 119-129 are helical when looking at residues V5-T132. ...
Embodiment 3
[0088] RGS4-G iαi The complex (AGR1) X-ray structure was read into SYBYL (Tripos) and all substructures except strand E (RGS4) were deleted. Also, remove all water. Polar hydrogen was added and Kollman United Atom force field optimization was used. All remaining hydrogen was then added. MOLCAD was then used to generate a surface of all residues believed to be involved in T182 (Ga-binding site) binding. These RGS4-core residues include: I21, I27, F30, F33, L34, E37, S39, N42, I43, W46, I110, L113, M114, D117, S118, R121. The surface area is calculated based on MOLCAD surfaces. The surface area of the same residues in the free RGS4 NMR structure was also calculated using MOLCAD. The calculated surface area of the free RGS4 NMR structure is 404.56 Å 2 . The calculated surface area of the crystal structure is 321.88 Å 2 . The difference in surface area is 82.67 Å 2 , that is about a 20% change in surface area between the two structures. The surface area of the M...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com