Primer sequence for cauliflower mosaic virus 35s promoter-containing transgenic crop nucleic acid amplification
A cauliflower flower leaf and primer sequence technology, applied to the determination/testing of microorganisms, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
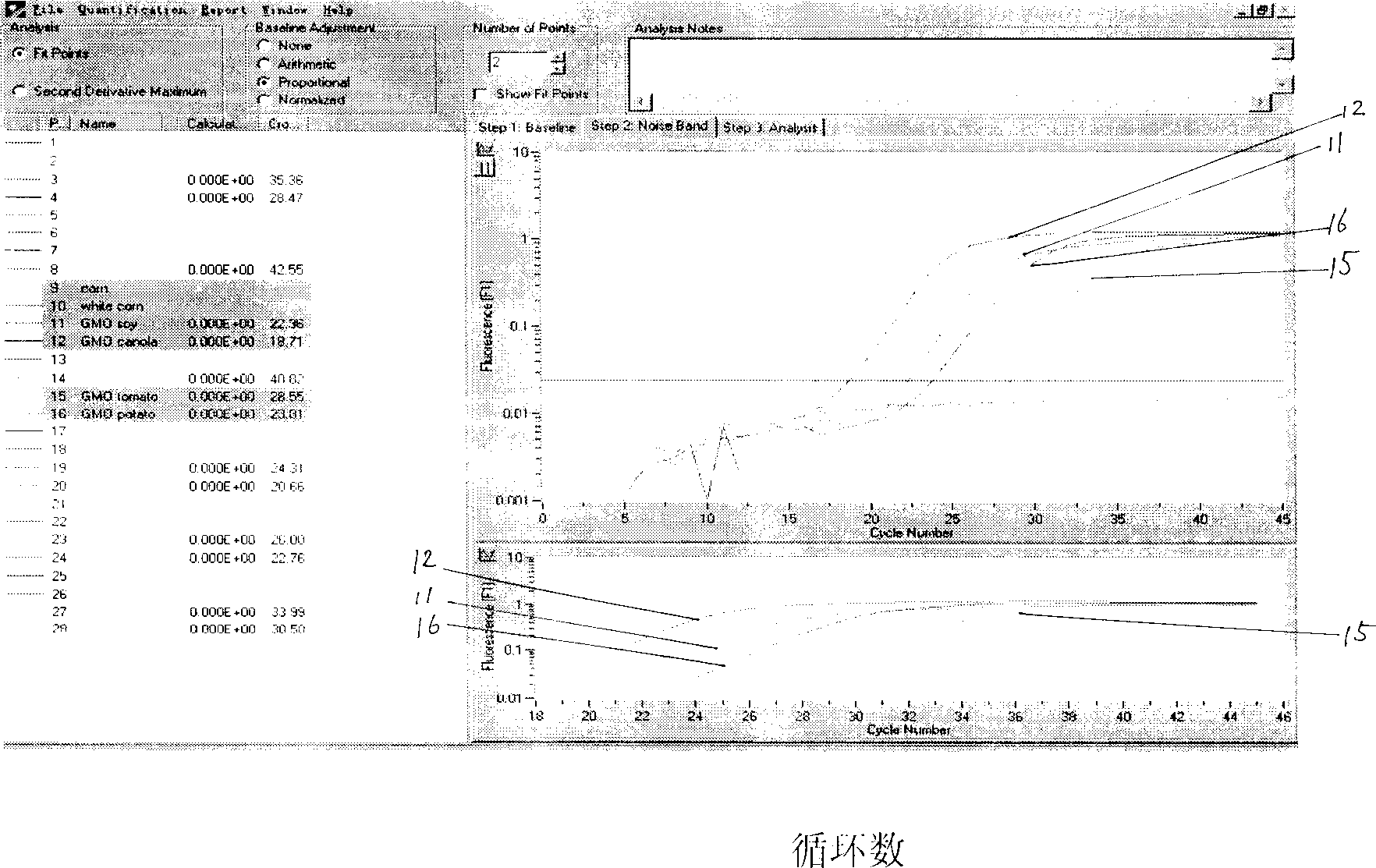
[0068] Qualitative detection: Select genetically modified and non-genetically modified agricultural products such as soybeans, corn, rape, tomato, and potatoes, and extract genomic DNA with the Promega magnetic bead method, specifically: weigh 100mg of plant material, place it in a 2ml centrifuge tube; add 500ul Lysis Buffer A and 5ul RNase A, mix with the material; add 250ulLysis Buffer B, mix well, and let stand at room temperature for 10 minutes; add 750ul precipitation buffer, mix well and centrifuge at high speed (13000×g) for 10 minutes; pipette the supernatant into a clean 2ml Add 50ul magnetic powder solution to the centrifuge tube, mix well; add 0.8 volume of isopropanol to the mixture, mix well, and let it stand at room temperature for 5 minutes; put the centrifuge tube on the magnetic rack for 1 minute, remove the clarified liquid; take out the centrifuge tube, and then Add 250ul Lysis Buffer B, mix well and place on the magnetic rack for 1 minute to remove the clari...
Embodiment 2
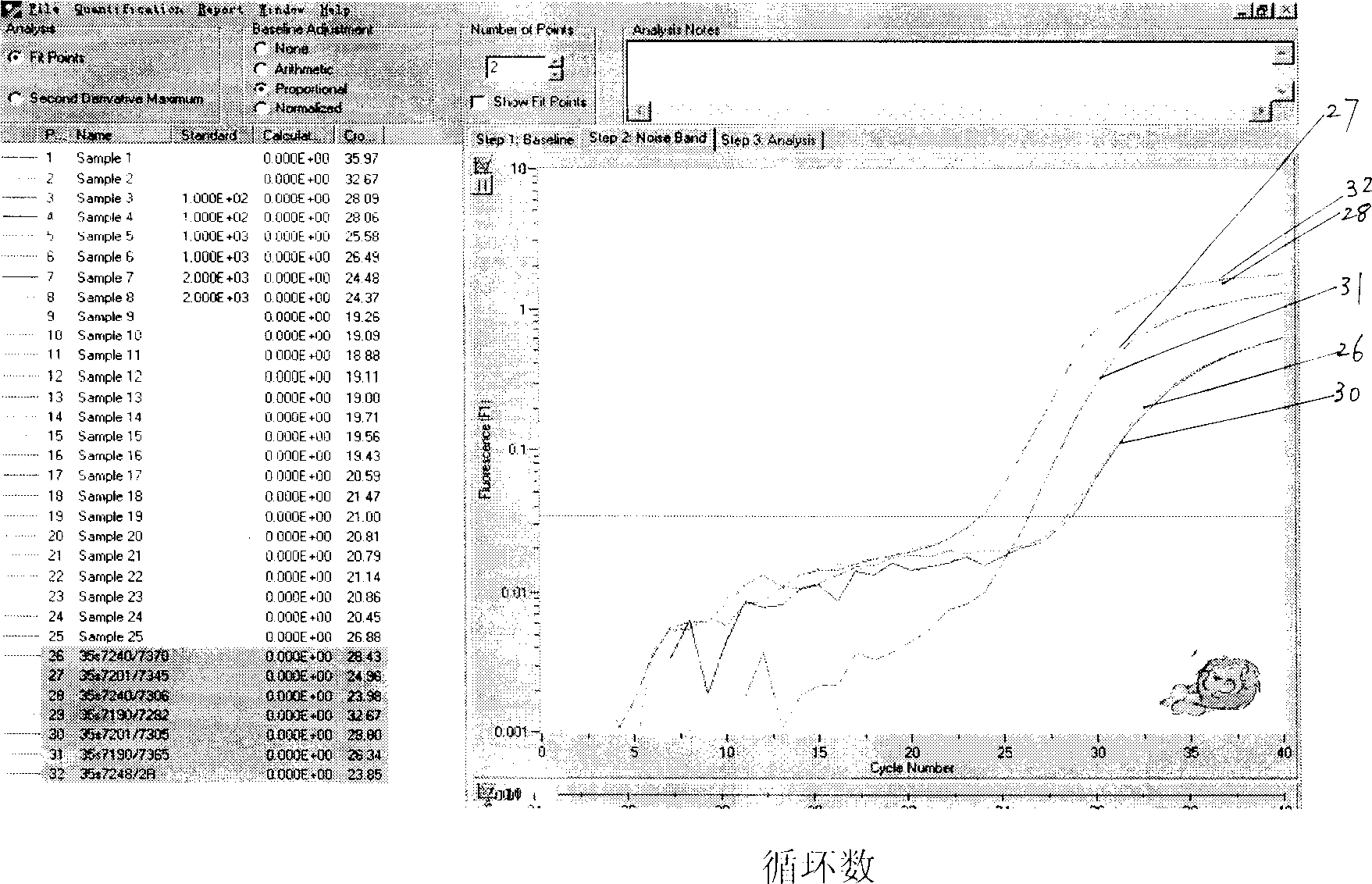
[0071] Quantitative detection: Take the Fluka soybean standard product that meets the international standard, the genomic DNA extraction method is the same as before, and use the 35s7240 / 35s2R primers for quantitative detection of fluorescent PCR, and the fluorescent PCR instrument uses RocheLight-Cycler. The results are shown in Table 4
[0072] sample
[0073] The regression curve that obtains from above-mentioned data is y=-0.346x+7.9383; Correlation r2=0.99; Then use 1.0% and 2.0% standard substance as tested sample, the GMO content that measures is respectively 0.94% and 2.26%, The quantitative deviation is 5%-15% (the quantitative deviation of GENESCAN company in the European Union is ≤20%), suggesting that the detection efficiency of the primers used has reached the international level of this project in the field of nucleic acid amplification detection.
Embodiment 3
[0075] Electrophoresis detection: take the Fluka transgenic soybean standard product that meets international standards, use the same method for genomic DNA extraction as before, and use 35s7240 / 35s2R primers for PCR amplification. The PCR amplification conditions are as follows:
[0076] 93°C: 3min 1 cycle
[0077] 93°C: 15 sec; 60°C: 30 sec; 72°C 1min 40 cycles
[0078] Take the above-mentioned amplification products, use 1% agarose, 2V / CM electrophoresis for one hour, soak in EB working solution for 15 minutes, and observe the results under ultraviolet light: a clear amplification band can be seen in positive samples, while negative samples have no amplification belt, see image 3 .
[0079] image 3 middle
[0080] 1) 0.0% Genetically Modified Soybean Standard 2) 0.1% Genetically Modified Soybean Standard
[0081] 3) 0.5% genetically modified soybean standard product 4) 2.0% genetically modified soybean standard product
[0082] M) 2000ladder
[0083] Advantages of th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com