Method for preparing oligonucleotide chip in use for DNA surface computer in heterozygosis with computer
An oligonucleotide and surface computing technology, which is applied in the field of preparation of oligonucleotide chips for DNA surface computing, can solve the problems of increasing variable operation, inability to use, errors, etc., and achieves simple operation process and reduced false positive rate. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] A DNA surface computing chip for solving SAT problems hybridized with computer computing
[0030] The present invention is an example with a SAT problem of 4 variables, and this problem is described as:
[0031] F=(w∨x∨y)∧(w∨y∨z)∧(x∨y)∧(w∨y)
[0032] Find the values of the four variables when F=1
[0033] The steps of DNA surface calculation chip design and hybrid calculation of this problem with electronic computer are described as follows:
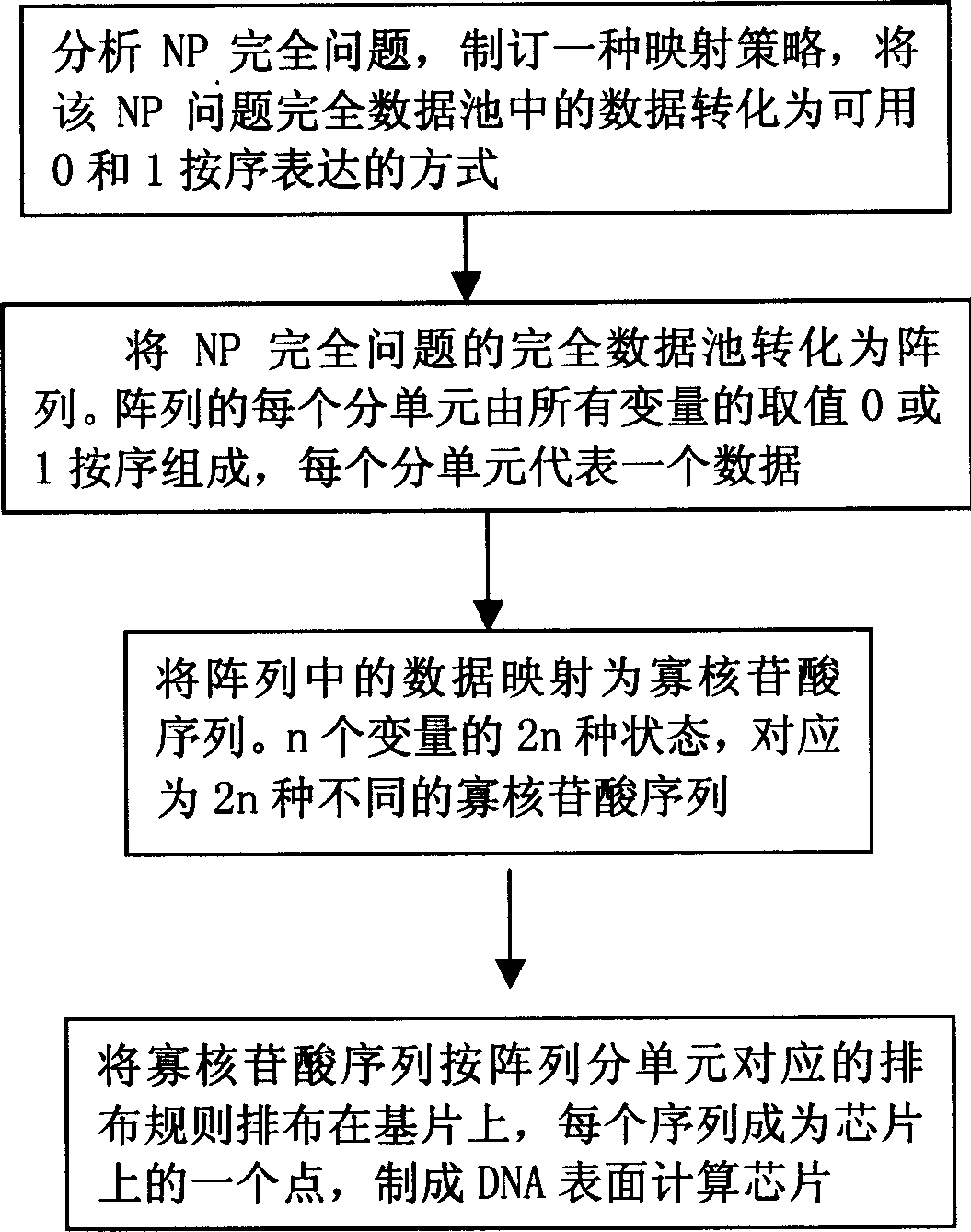
[0034] (1) Generate a DNA surface computing chip
[0035] ① The SAT question contains 4 variables (w, x, y, z) and 4 clauses, each variable takes a value of 1 or 0, then the complete data pool of the question contains 2 4 = 16 data, arranged in the order of wxyz, the 16 data are as follows: 0000, 0001, 0010, 0011, 0100, 0101, 0110, 0111, 1000, 1001, 1010, 1011, 1100, 1101, 1110, 1111
[0036] x
[0037] 0 0
[0038] variable value
[0039] The oligonucleotide chain encoded by the above SAT pr...
Embodiment 2
[0048] DNA Surface Computing Chip for Solving the Maximum Clique Problem Hybridized with Computer Computing
[0049] A group of a graph G is a vertex subset S of the graph G, so that there is an edge connection between any two vertices in S. If there is |S|≥|S′| for any other group S′ of G, it is called S is the largest clique of G. The maximum clique problem of a graph is an NP-complete problem.
[0050] The present invention takes a given graph with 6 vertices as an example. The given figure is attached Figure 3a , manually draw its complement as attached Figure 3b .
[0051] The steps of DNA surface calculation chip design and hybrid calculation of this problem with electronic computer are described as follows:
[0052] (1) Generate a DNA surface computing chip
[0053] ① The maximum clique problem contains 6 vertices. First, define a mapping strategy for a complete data pool:
[0054] a. Vertices are represented by A1, A2, A3, A4, A5, and A6 respectively.
[0055...
Embodiment 3
[0073] A DNA surface computing chip for solving the maximum vertex segmentation problem of graphs hybridized with computer computing. The maximum vertex segmentation problem of a graph is, given a graph G, there is a subset S of vertices in graph G, and the total number of connecting lines between any vertex in S and vertices outside the subset is the largest, then this vertex The subset is the maximum vertex split for a given graph G. This is also an NP-complete problem.
[0074] The present invention takes a given graph with 6 vertices as an example, and the given graph is as attached Figure 5 .
[0075] The model and steps of the DNA computing chip to calculate this problem are as follows:
[0076] (1) Generate a DNA surface computing chip
[0077] ① The maximum vertex segmentation problem contains 6 vertices. First, define a mapping strategy for a complete data pool:
[0078] a There are two assignments for setting Ai, when the vertex is in the vertex subset, the val...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com