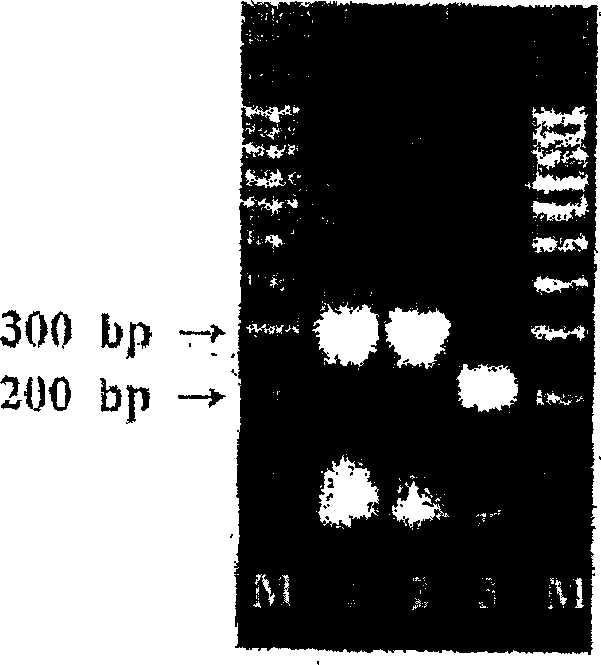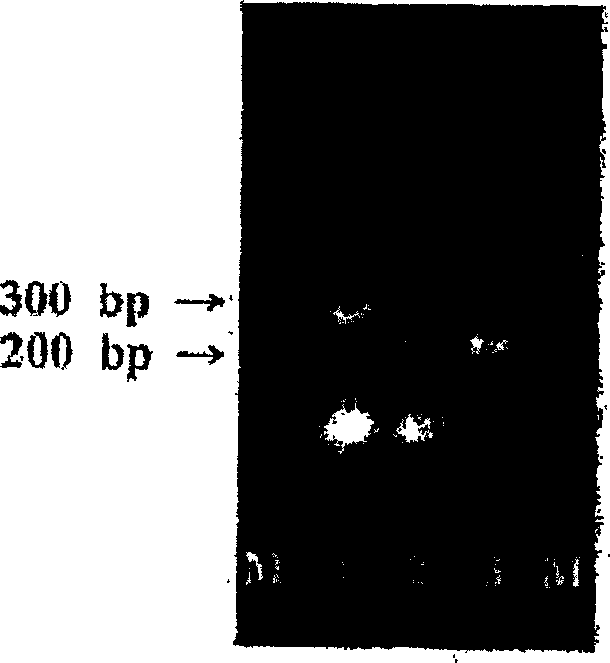General primers and process for detecting diverse genotypes of human papillomavirus by PCR
A technology for human papilloma and universal primers, applied in biochemical equipment and methods, microbiological determination/inspection, etc., can solve the problems of HPV lack of sensitivity, limited use, and primer set defects, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1: PCR primer design and synthesis
[0050] (1) PCR primer design
[0051] According to the purpose of the present invention, a nucleotide sequence that can complementarily bind to different types of HPV and thereby amplify all HPV type DNA is designed as a universal primer for PCR.
[0052] First, export HPV 1a, 2a, 3, 4, 5, 6b, 7, 8, 9, 10, 11, 12, 13, 15, etc. from the NCBI (National Center for Biotechnology Information) and the National HPV database of Los Alamos. 16, 16r, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35h, 36, 37, 38, 39, 40, 41, 42, 44, 45, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 63, 65, 66, 67, 68, Full-length nucleic acid sequences of 72 HPV types 70, 72, 73, 74, 75, 76, 77 and 80.
[0053] Using Clustal W computer program (DNSSTAR, MegAlign TM 5. DNASTAR Inc.) conducts DNA sequence homology research on the derived DNA sequence. This program screens representative base sequences in the group thr...
Embodiment 2
[0065] Example 2: PCR primer design and synthesis
[0066] Except that the designed universal PCR primers are set to about 24±3 bp and about 33±2 bp nucleotide sequences, and their amplified products are about 170-550 bp, Example 2 is completed under the same conditions and steps as Example 1.
[0067] Under the above conditions, the final 8 pairs of universal primers were selected. For ease of description in the following, each of the 8 pairs of combination primers according to the embodiment of the present invention is referred to as AlbioGP3, AlbioGP4, AlbioGP5, AlbioGP6, AlbioGP7, AlbioGP8, AlbioGP9, AlbioGP10, and AlbioGP11, respectively. Table 3 shows the nucleic acid sequence, SEQ ID No, the type of amplified HPV, the size of the amplified product, and the actual amplification result of the amplified region of the determined primer pair. And Table 4 shows the actual amplification product size of representative cervical cancer-related HPV types by using AlbioGP5, AlbioGP6, ...
Embodiment 3
[0075] Example 3: PCR amplification using universal primers
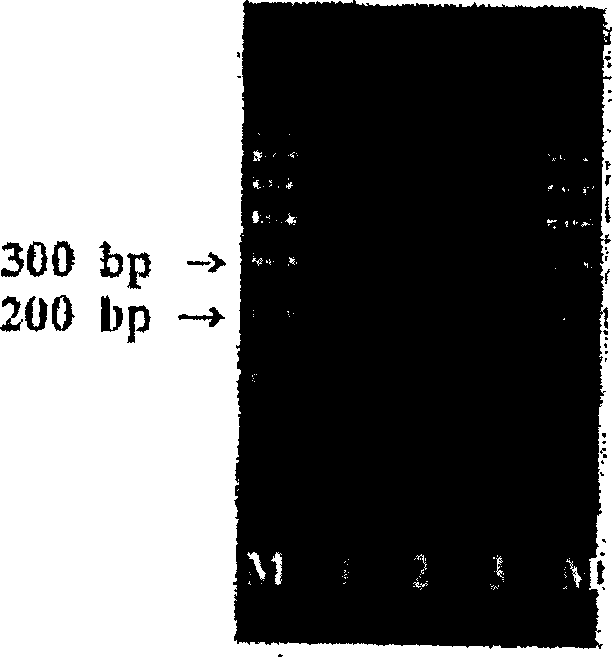
[0076] The 3 pairs of primers (AlbioGP1, AlbioGP2 and AlbioGP3) synthesized in Example 1 were used to amplify the target HPV 16 infected Caski cell line (ATCC CRL-1550), HPV 18 infected HeLa cell line (ATCC CCL-2) and Non-HPV infected K-562 cell line (KCLB-10243, Korean cell line bank).
[0077] Culture each of Caski, HeLa, and K-562 cells according to the manufacturer’s instructions, then add 0.25% trypsin solution to Caski and HeLa cells, and centrifuge the cultured cell lines from the culture flask. Wash each cultured Caski, HeLa and K-562 cell lines once with Dulbecco’s phosphate buffered saline (Gibco Inc.), and check the number of cells under a microscope. Then the 2×10 from the cultured cell line 4 The cells were suspended in 230 μL (microliter) of D.W and heated at 100°C (Celsius) for 15 minutes to extract cellular DNA. The extracted cellular DNA was cooled at room temperature and centrifuged at 12,000 rpm for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com