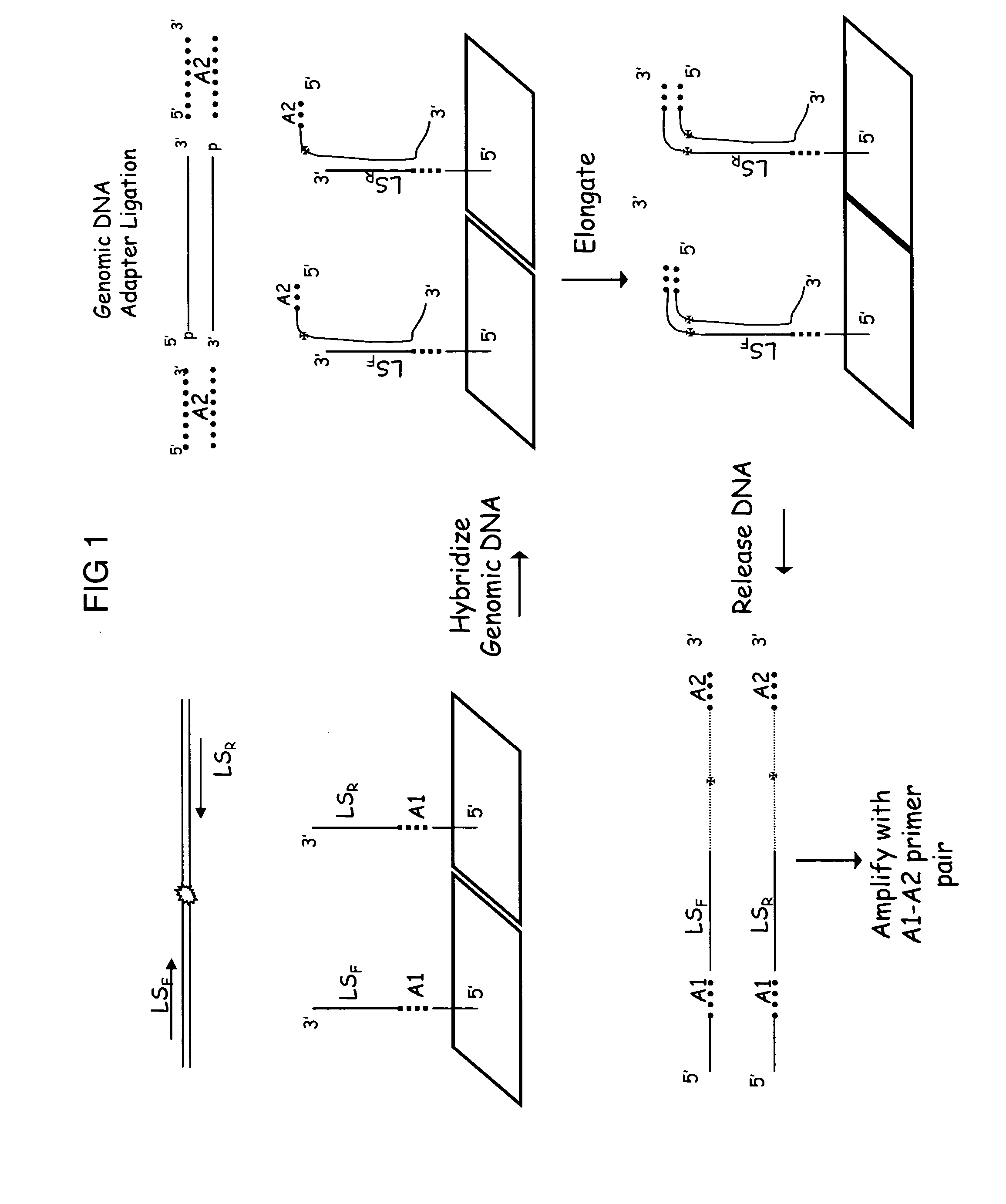
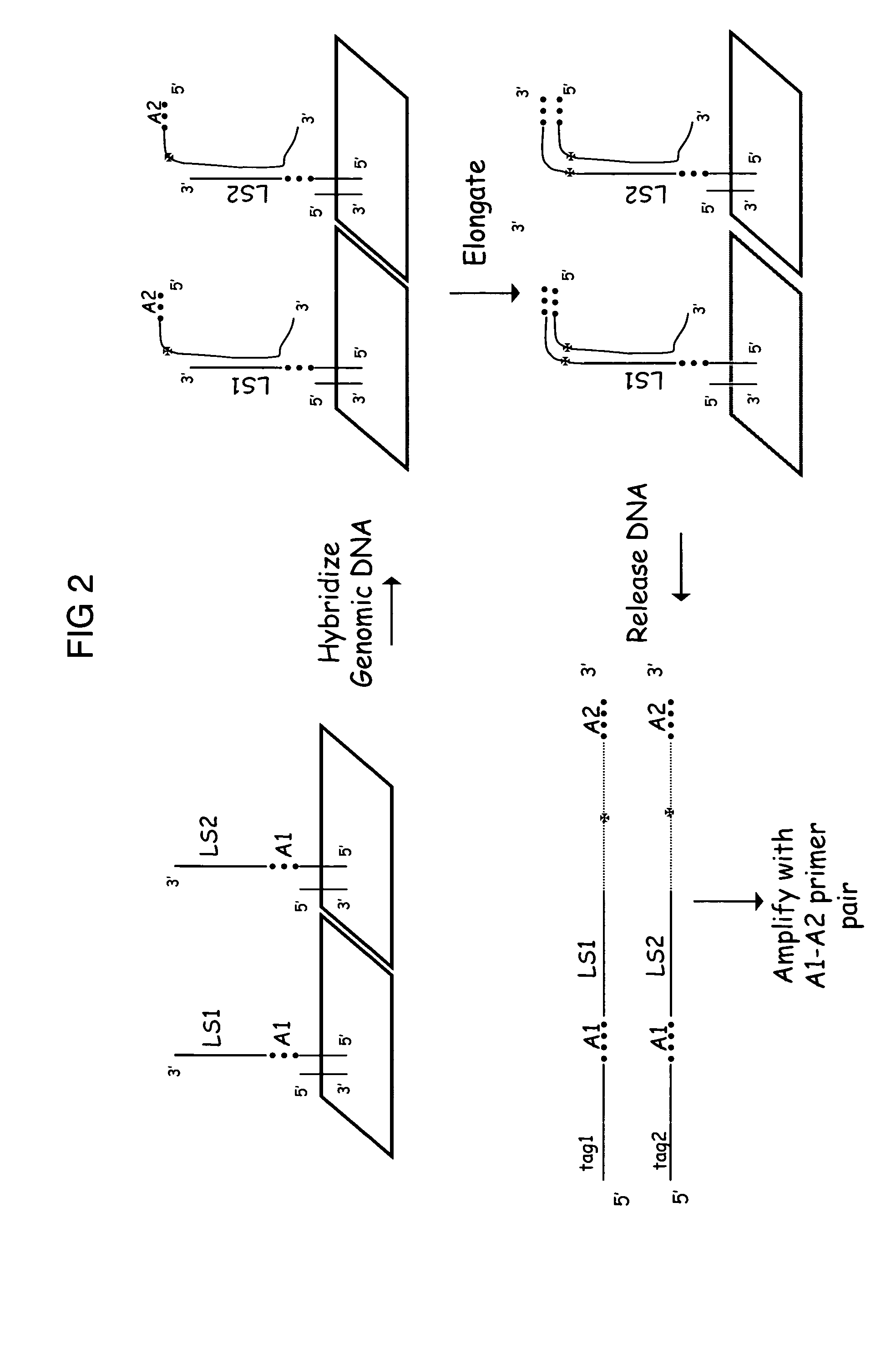
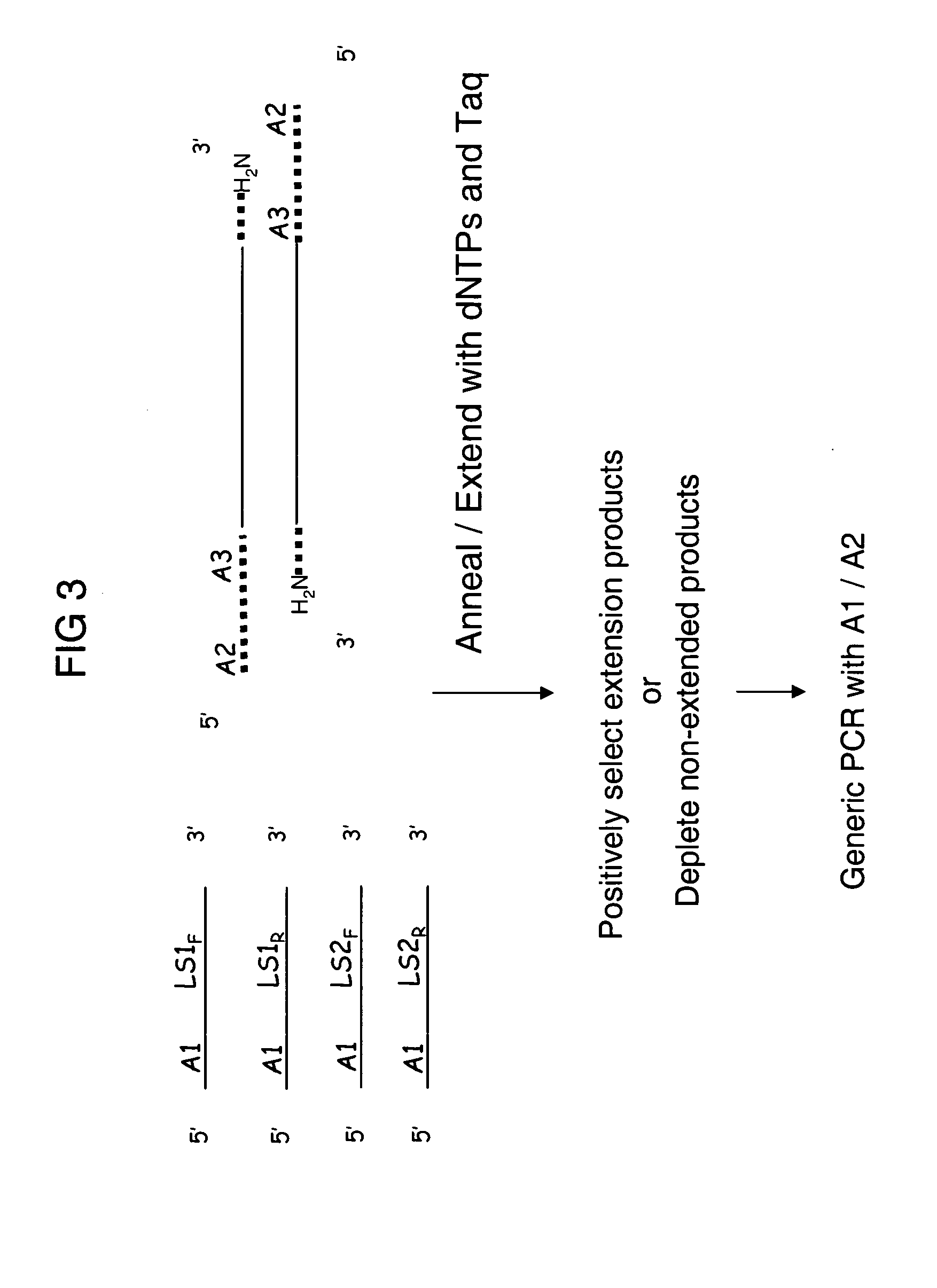
Complexity management of genomic DNA by locus specific amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Multiplexed Anchored Runoff Amplification
[0119] Genomic DNA was digested with MseI and ligated to an adapter containing T7 promoter sequence as a priming site. The final concentration of the genomic DNA was 10 ng / μl in 1×T4 DNA Ligase Buffer. To generate extended capture probes 2.5 μl of adapter ligated DNA, 2.5 μl 10×Taq Gold Buffer, 2 μl 25 mM MgCl2, 2.5 μl 10×dNTPs, 5 μl of a 500 nM mixture of 150 different capture probes in TE buffer corresponding to 150 different forward primers from the HuSNP assay, 0.25 μl Perfect Match Enhancer, 0.25 μl AmpliTaq Gold (Applied Biosystems, Foster City, Calif.) and 10 μl of water were mixed to give a final reaction volume of 25 μl. The reaction was incubated at 95° C. for 6 min followed by 26 cycles of 95° C. for 30 sec, 68° C. for 2.5 min (decreasing 0.5° C. on each subsequent cycle) and 72° C. for 1 min, then to 4° C.
[0120] The extended capture probes were made double stranded by the addition of 0.25 μl of 1 μM T7 primer and incubation at ...
example 2
Multiplexed Anchored Runoff Amplification with Biotin Enrichment
[0123] Prepare adaptor ligated genomic DNA as above. To generate extended capture probes 2.5 μl of adapter ligated DNA, 2.5 μl 10×Taq Gold Buffer, 2 μl 25 mM MgCl2, 0.5 μl 50×acGT (6 mM dATP, 6 mM dCTP, 10 mM dGTP, 10 mM dTTP), 5 μl of a 500 nM mixture of 150 different capture probes in TE buffer corresponding to 150 different forward primers from the HuSNP assay, 0.25 μl Perfect Match Enhancer, 0.25 μl Amplitaq Gold, 2 μl 1 mM Biotin-N6-dATP (Perkin Elmer, Boston, Mass.), 2 μl 1 mM Biotin-N4-dCTP (Perkin Elmer) and 8 μl of water were mixed to give a final reaction volume of 25 μl. The reaction was incubated at 95° C. for 6 min followed by 26 cycles of 95° C. for 30 sec, 68° C. for 2.5 min (decreasing 0.5° C. on each subsequent cycle) and 72° C. for 1 min, then to 4° C. Pass reaction over G-25 Sephadex column to remove unincorporated biotin-dNTPs.
[0124] Enrich for biotinylated extension products. Adjust the G-25 elua...
example 3
Multiplexed Anchored Runoff Amplification with Exo III Enrichment.
[0127] Prepare adaptor ligated genomic DNA as above. Kinase capture probes by incubating 12 μl of a 150-plex stock of either forward or reverse HuSNP® primers with 12.7 μl H2O, 3 μl 10×T4 polynucleotide kinase buffer, 0.3 μl 100 mM ATP, and 2 μl T4 Polynucleotide Kinase. Incubate the reaction at 37° C. for 30 min. Adjust reaction volume to 50 μl and pass reaction over G-25 column to exchange buffer.
[0128] To generate extended capture probes 5 μl of adapter ligated DNA, 5 μl 10×Taq Gold Buffer, 4 μl 25 mM MgCl2, 5 μl 10×dNTPs, 20 μl of the kinased mixture of 150 different capture probes, 1 μl Perfect Match Enhancer, 0.5 μl AmpliTaq Gold and 9.5 μl of water were mixed to give a final reaction volume of 50 μl. The reaction was incubated at 95° C. for 6 min followed by 26 cycles of 95° C. for 30 sec, 68° C. for 2.5 min (decreasing 0.5° C. on each subsequent cycle) and 72° C. for 1 min, then finally to 4° C. Pass the re...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Electrical resistance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com