RNA interference pathway genes as tools for targeted genetic interference
a technology of rna interference pathway and gene, applied in the field of genes, can solve the problems of more undesirable effects of “cosuppression” and transgene silencing, and achieve the effects of promoting dsrna-mediated genetic interference, reducing or increasing the activity of the rnai pathway
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Strains and Alleles
[0127] The Bristol strain N2 was used as standard wild-type strain. The marker mutations and deficiencies used are listed by chromosomes as follows: LGI: dpy-14(e188), unc-13(e51); LGIII: dpy-17(e164), unc-32(e189); LGV: dpy-11(e224), unc-42(e270), daf-11(m87), eDf1, mDf3, nDf31, sDf29, sDf35, unc-76(e911). The C. elegans strain DP13 was used to generate hybrids for STS linkage-mapping (Williams et al., 1992, Genetics 131:609-624).
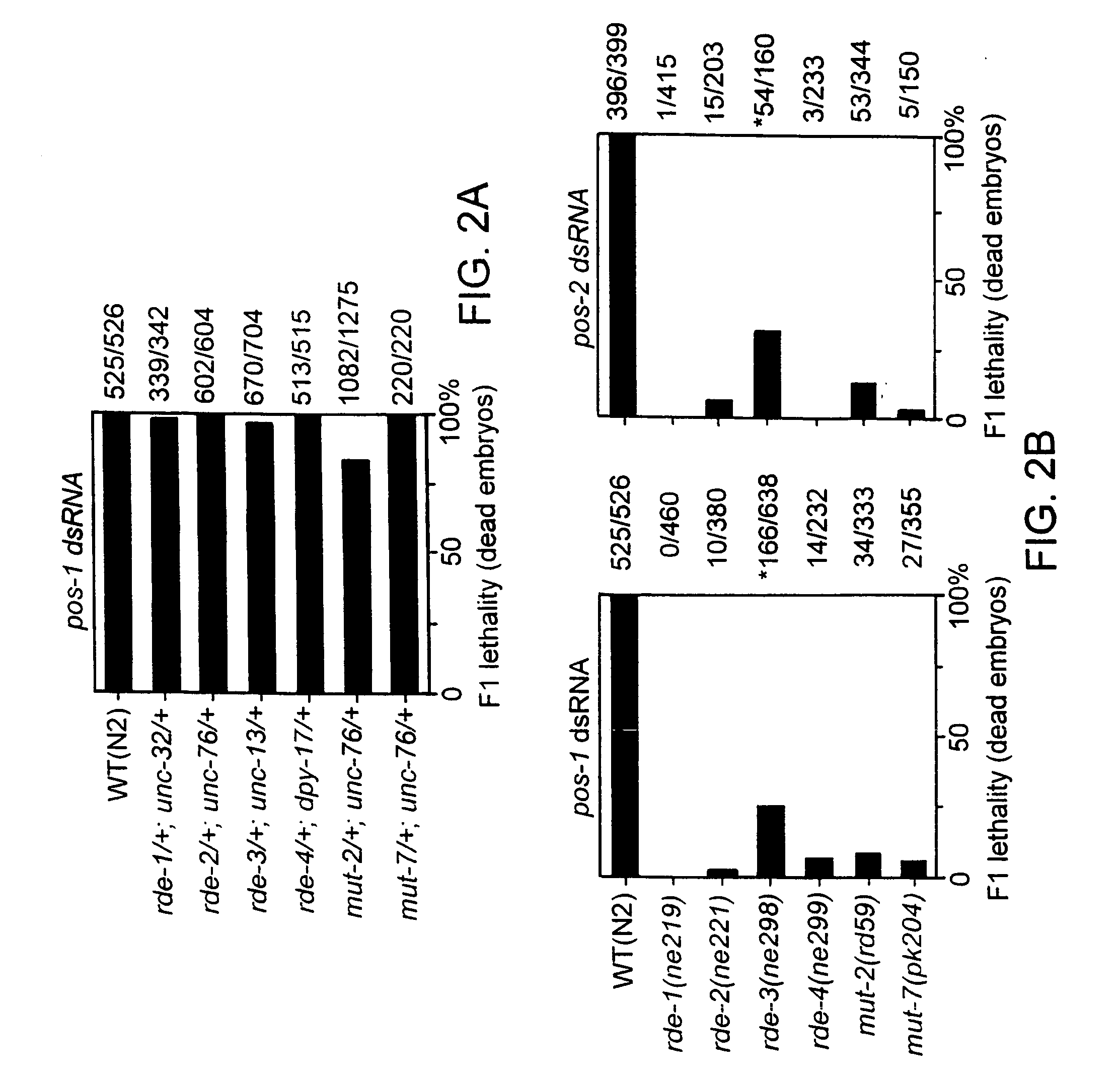
[0128] Sensitivity to RNAi was tested in the following strains. MT3126: mut-2(r459) (obtained from John Collins, Department of Biochemistry & Molecular Biology, University of New Hampshire, Durham, N. H.); dpy-19(n1347), TW410: mut-2([459) sem-4(n1378), NL917: mut-7(pk204), SS552: mes-2(bn76) rol-1(e91) / mnC1 (obtained from S. Strome, Biology Dept., Indiana University), SS449: mes-3(bn88) dpy-5(e61) (from S. Strome, supra); hDp20, SS268: dpy-11(e224) mes-4(bn23) unc-76(e91) / nT1, SS360: mes-6(bn66) dpy-20(e1282) / nT1, CB879: him-1 (e879)....
example 2
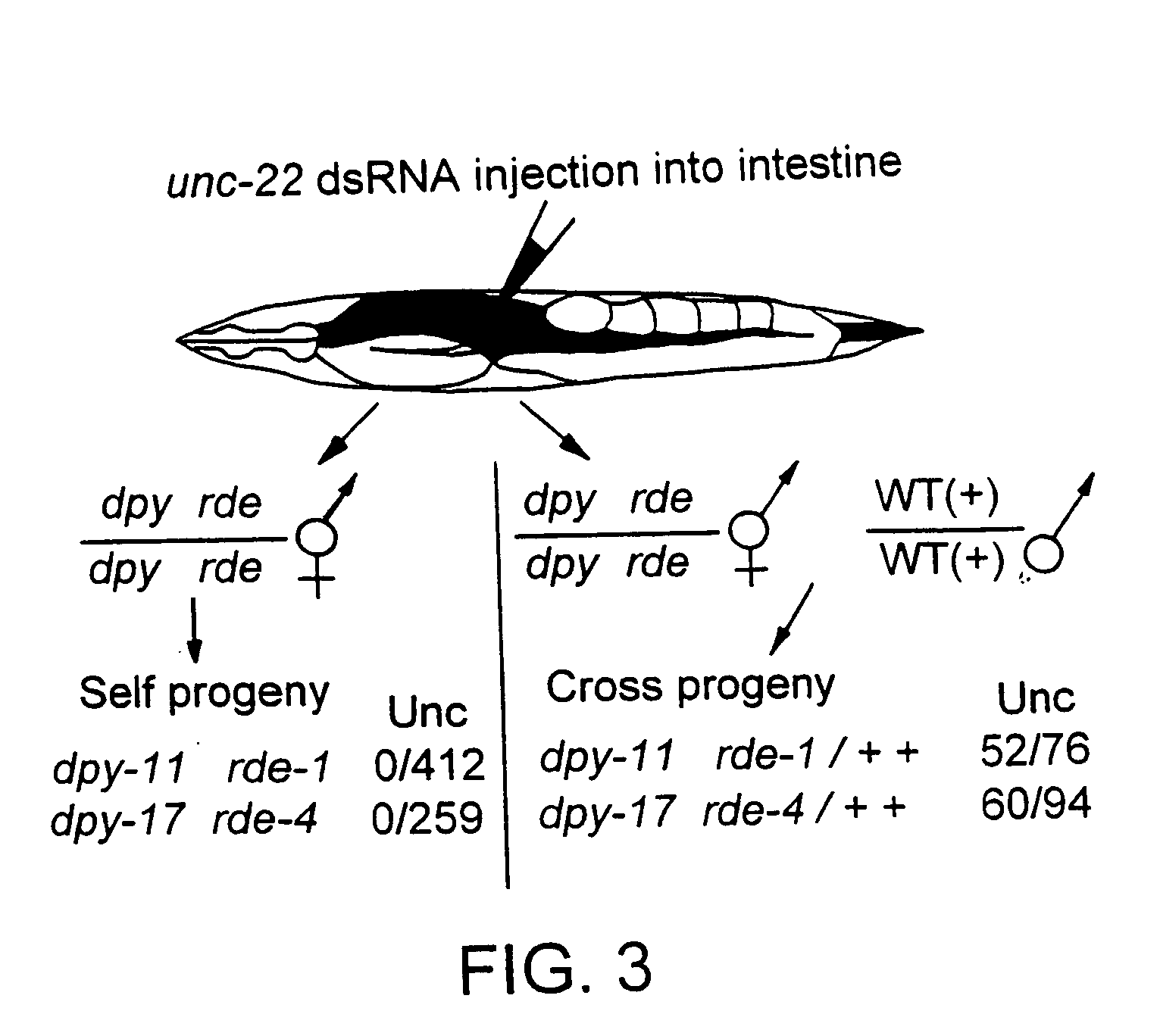
[0131] Genetic interference using RNAi administered by microinjection was performed as described in Fire et al., 1998, supra and Rocheleau et al., 1997, Cell 90:707-716. pos-1 cDNA clone yk61h1, par-2 cDNA clone yk96h7, sqt-3 cDNA clone yk75f2 were used to prepare dsRNA in vitro. These cDNA clones were obtained from the C. elegans cDNA project (Y. Kohara, Gene Network Lab, National Institute of Genetics, Mishima 411, Japan).
[0132] Genetic interference using RNAi administered by feeding was performed as described in Timmons and Fire, 1998, Nature 395:854. pos-1 cDNA was cloned into a plasmid that contains two T7 promoter sequences arranged in head-to-head configuration. The plasmid was transformed into an E. coli strain, BL21 (DE3), and the transformed bacteria were seeded on NGM (nematode growth medium) plates containing 60 μg / ml ampicillin and 80 μg / ml IPTG. The bacteria were grown overnight at room temperature to induce pos-1 dsRNA. Seeded plates (BL21(DE3)...
example 3
Identification of RNAi-Deficient Mutants
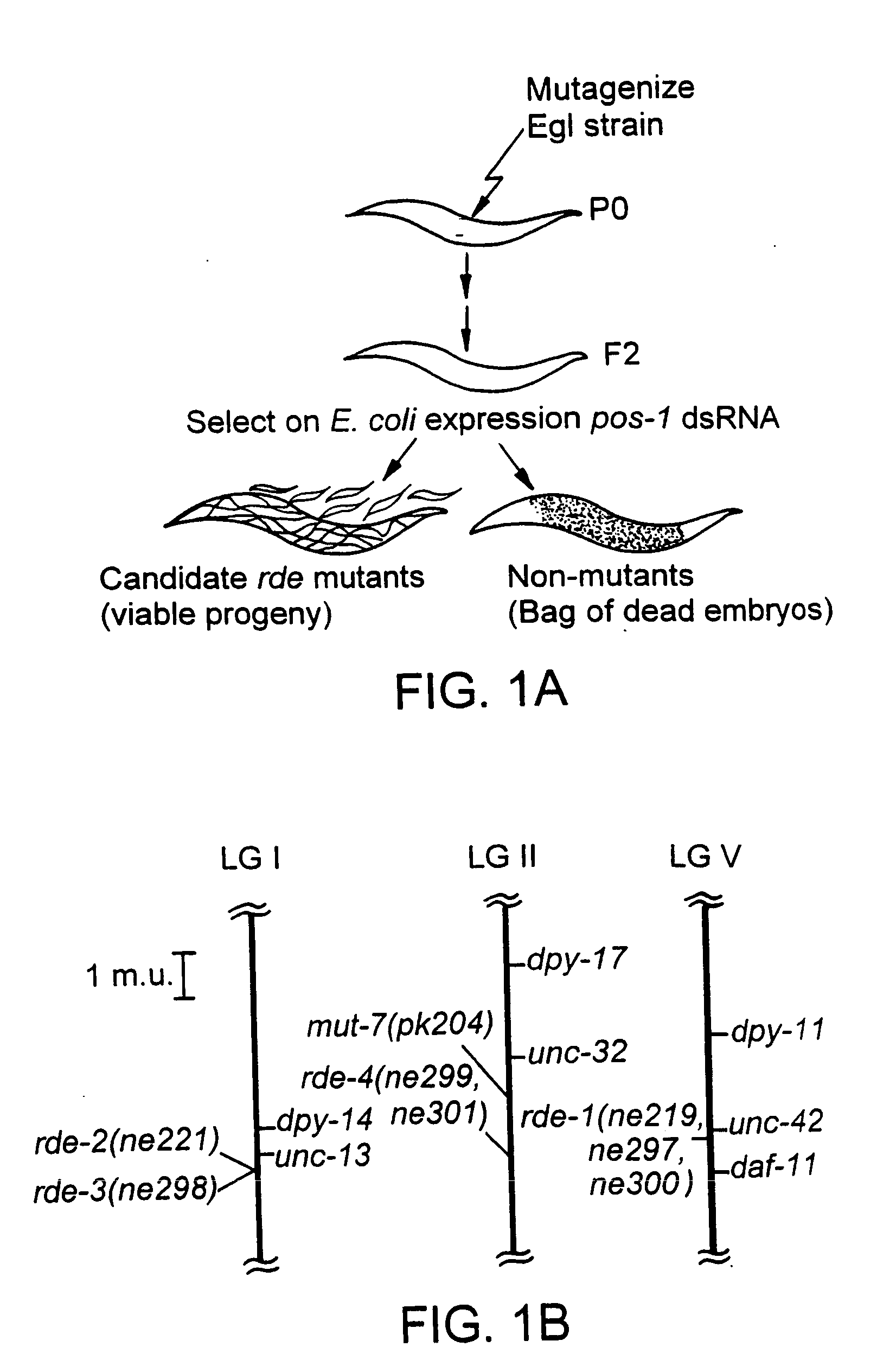
[0134] A method of screening for mutants defective in the RNAi pathway was devised that would permit the large-scale application of dsRNA to mutagenized populations. Feeding worms E. coli which express a dsRNA, or simply soaking worms in dsRNA solution, are both sufficient to induce interference in C. elegans (Timmons and Fire, 1998, supra; Tabara et al., 1998, Science 282:430-431). To carry out a selection, the feeding method was optimized to deliver interfering RNA for an essential gene, pos-1. C. elegans hermaphrodites that ingest bacteria expressing dsRNA corresponding to a segment of pos-1 are themselves unaffected but produce dead embryos with the distinctive pos-1 embryonic lethal phenotype.
[0135] To identify strains defective in the RNAi pathway, wild-type animals were mutagenized, backcrossed, and the F2 generation examined for rare individuals that were able to produce complete broods of viable progeny. Chemical mutagenesis was use...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com