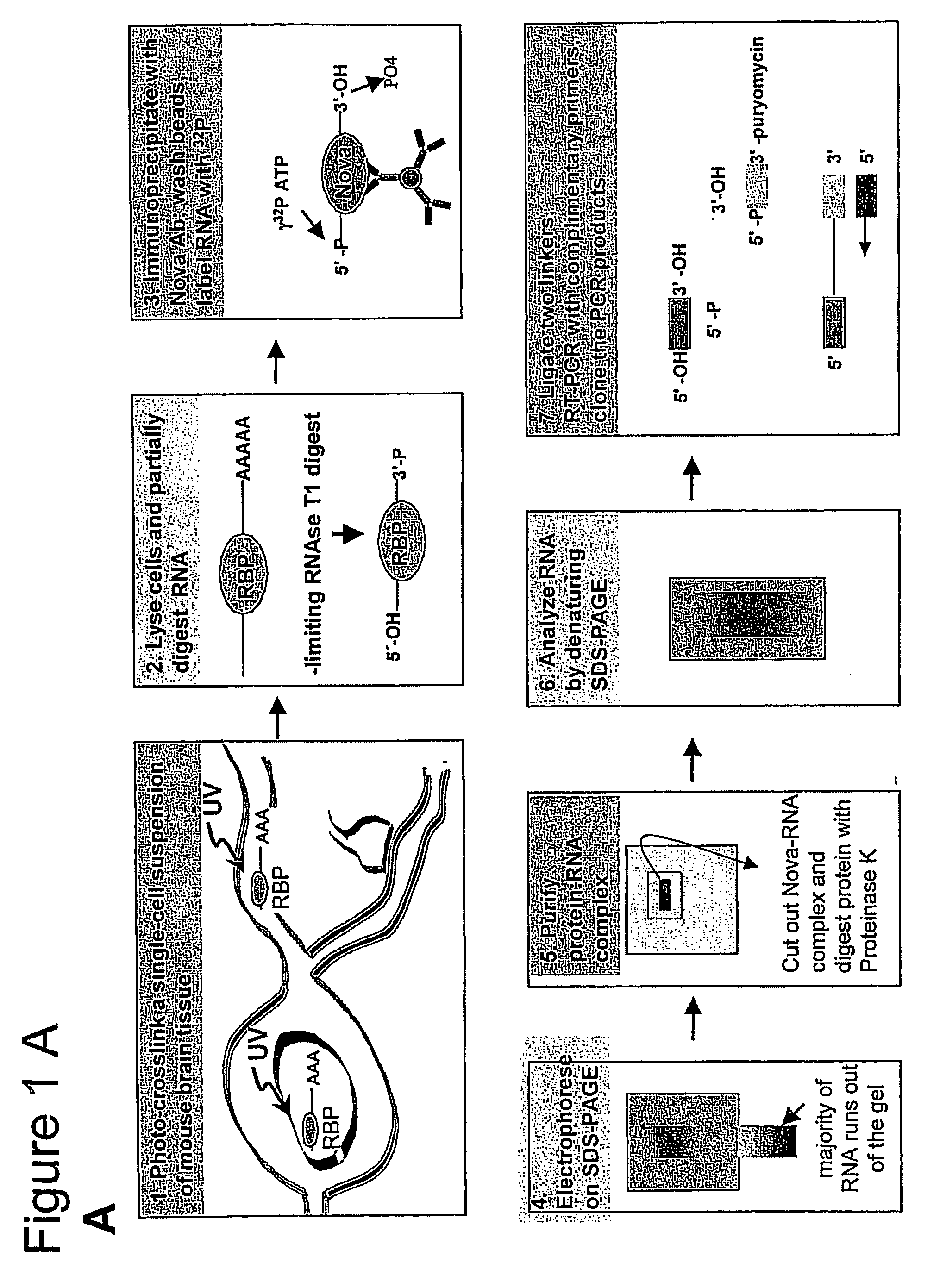
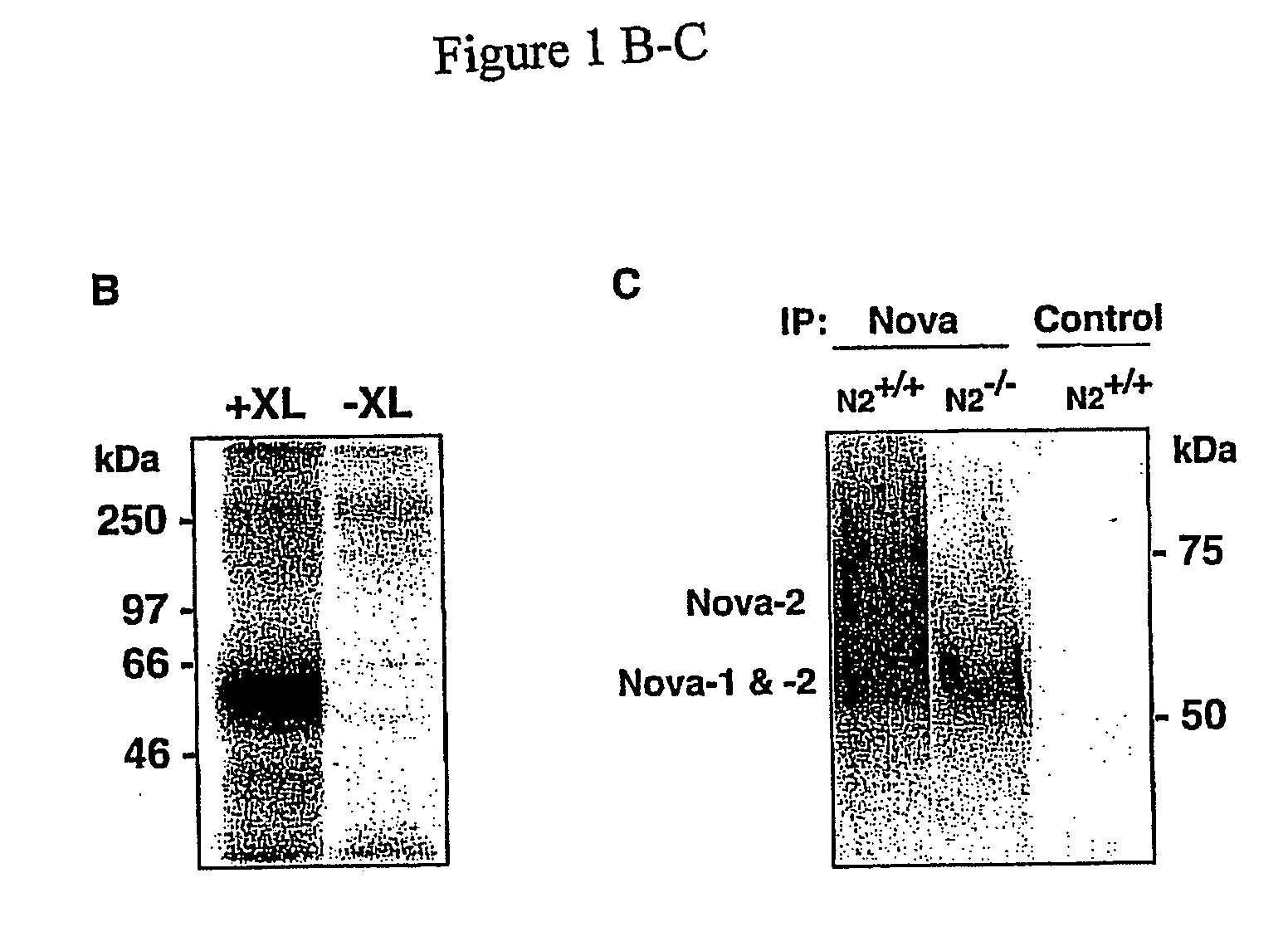
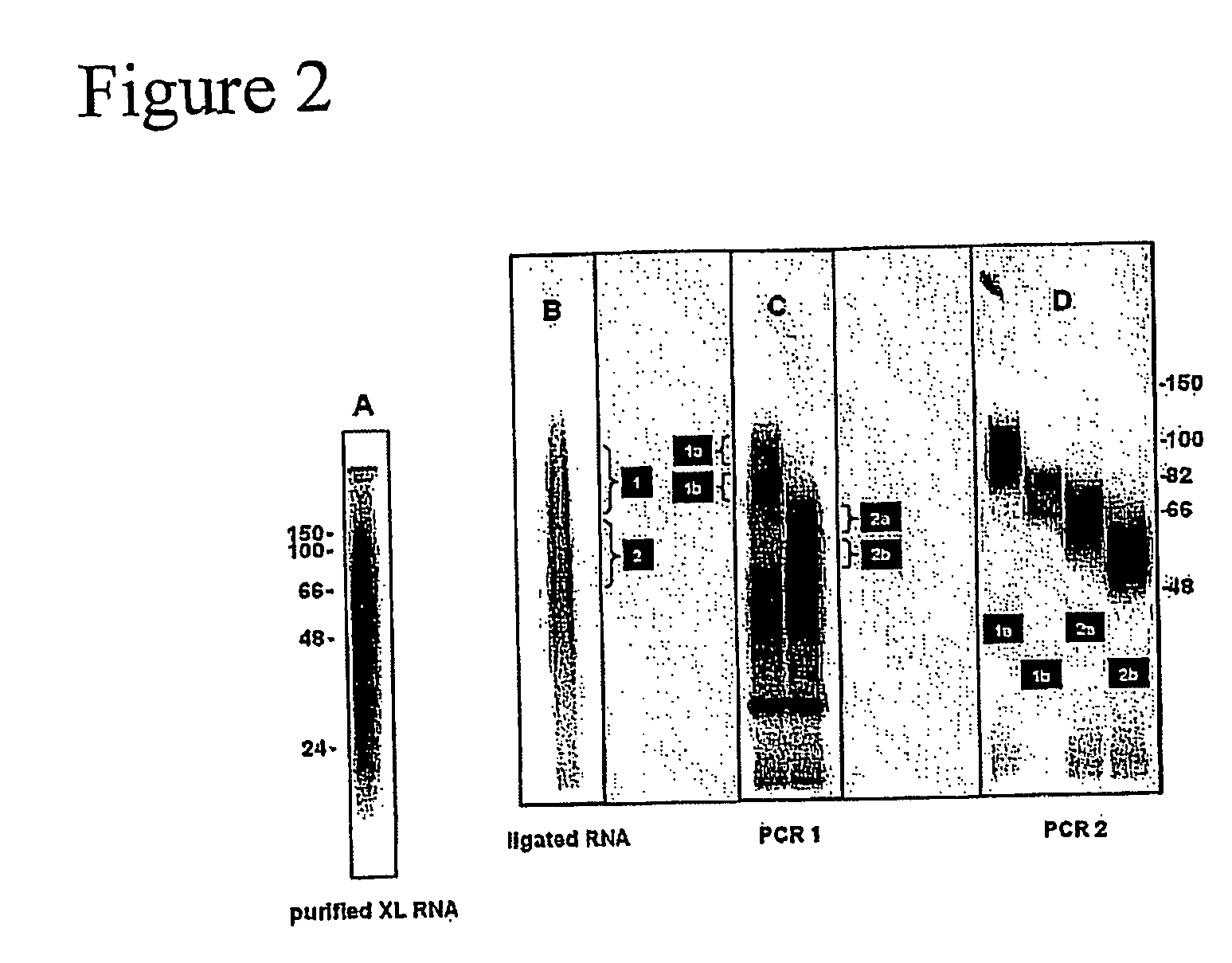
Method of purifying RNA binding protein-RNA complexes
a technology of rna binding protein and complex, which is applied in the field of purifying rna molecules, can solve the problems of difficult identification of rbps targets in a number of autoimmune and genetic diseases
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
CLIP Specifically Isolates Covalently Bound RBP-RNA Complexes
Materials and Experimental Methods
Immunoblot Analysis.
[0307] The following antibodies were used: gephyrin (Transduction laboratories), rabbit Nova antiserum (Buckanovich, R. J. et al, Mol Cell Biol 17, 3194), Hsp90 (Transduction laboratories), rabbit brPTB antiserum (Polydorides, A. D. et al, Proc Natl Acad Sci USA 97, 6350), dimethyl-Histone H3 (Upstate Biotechnology). Antibody to neuronal Hu proteins and Human POMA serum were obtained from paraneoplastic neurologic disease patients. BRPTB antibody was used as previously described (Polydorides AD et al, Proc Natl Acad Sci USA 97: 6350-55). Nova-1 knockout mice.
[0308] Nova-1 knockout mice were previously described (Jensen K B et al, Neuron 25:359-71).
UV Cross-Linking of Mouse Tissue.
[0309] Mouse hindbrain and spinal cord tissue was dissected from 60 postnatal day 4 P8 mice, and rapidly disaggregated in 50-milliliter (ml) polypropylene tubes using a rubber syringe p...
example 2
Monitoring of Linker Ligation and RT-PCR of RNA Molecules
[0333]32P-labeled RNA was size purified after cross-linking and purification from N2A cells using anti-Nova antiserum as described in Example 1. The size of the RNA fragments ranged from 24-150 bases; the modal size of the RNA was approximately 60 bases (FIG. 2A). Purified RNA fragments were ligated to 5′ and 3′ linker oligonucleotides (“linkers”), which added 16 bases to each end of the molecule. The majority of the labeled fragment RNA shifted in size by 32 bases, indicating successful ligation (FIG. 2B). RNA isolated from regions 1 and 2 in FIG. 2B was amplified by RT-PCR with specific primers complementary to the linker sequences (FIG. 2C). The prominent band at 32 bases was the product from the ligation of the two RNA oligonucleotides without insert. The products in (C) were further divided and further amplified by PCR (FIG. 2D).
example 3
CLIP Method using Nova-1 Antisera Enabled the Isolation and Identification of Sequence Fragments Containing Nova-1 Binding Sites
Materials and Experimental Methods
Computer Analysis of Nova CLIP Fragments.
[0334] 3400 control fragments were randomly generated by a computer program from a 200,000 nucleotides long sequence consisting of 66% intronic, 14% exonic and 20% 3′UTR sequences (corresponding to the ratio in Nova CLIP fragments) from random genes on mouse chromosome 1, such that they corresponded in their size to Nova CLIP fragments (with the average size of 71 nucleotides). Another program was made to count the number of particular polynucleotide (up to 20 nucleotides in a row) in each fragment, and calculate the frequency of fragments carrying a certain number of that polynucleotide (for example, YCAY, where Y represents either U or C). An additional program was made to calculate the average frequencies of nucleotides at three positions flanking a particular dinucleotide (CA...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Digital information | aaaaa | aaaaa |
| Gene expression profile | aaaaa | aaaaa |
| Ionicity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com