Methods and systems for monitoring production of a target protein in a nanolipoprotein particle
a technology of nanolipoproteins and target proteins, applied in the field of membrane associated proteins, can solve the problem of challenging study of protein classes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Cell-Free Production of NLPs bR Protein
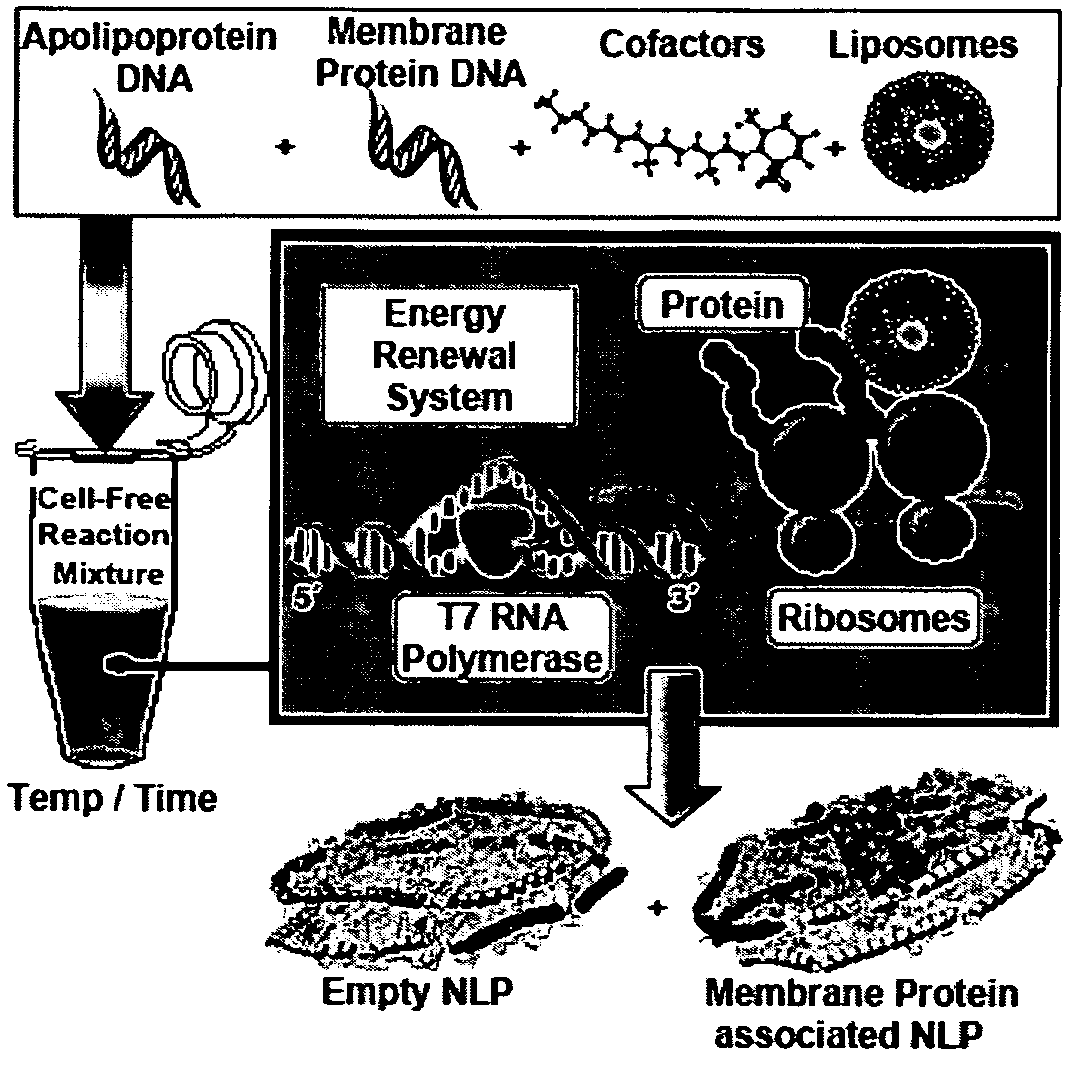
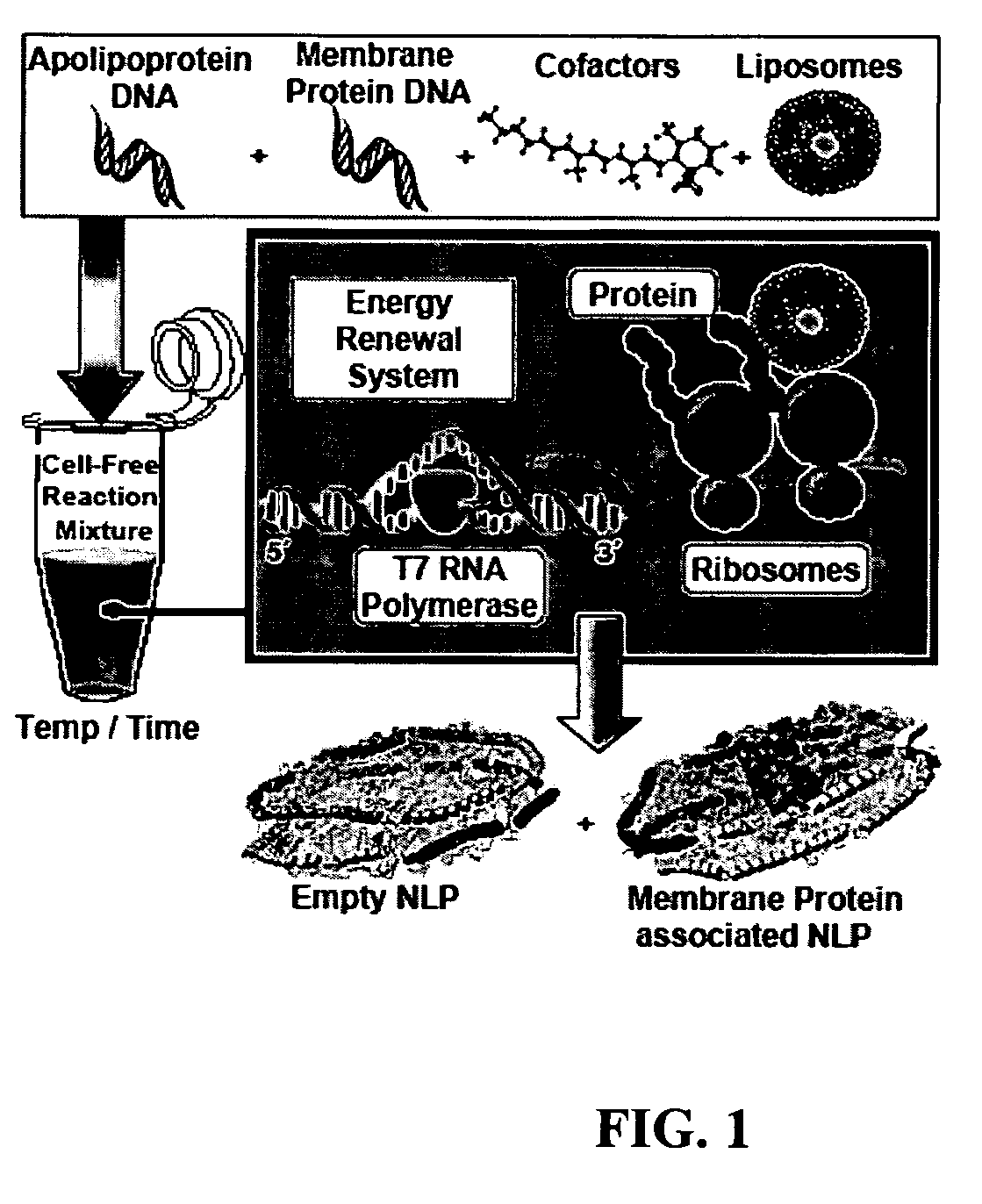
[0124]The experimental strategy for cell-free membrane protein-NLP self-assembly was based on the ability of membrane proteins to insert into lipid bilayers during cell-free synthesis, the apolipoprotein ability to sequester lipid bilayer patches, and the demonstrated ability of NLPs to than solubilize membrane proteins. Individual plasmid DNAs encoding the membrane protein and the apolipoprotein are added to the cell-free reaction with the addition of phospholipids and cofactors to produce membrane protein associated discoidal nanolipoprotein particles (NLPs) in a single reaction. In particular, as shown in FIG. 1 constituents (DNA, lipid vesicles, cofactors and cell-free lysates) are added together in a single reaction vial. The cell-free lysates take advantage of the T7 coupled transcription and translation system to produce a mixed population of self-assembled NLPs with and without associated integral membrane protein.
[0125]In a first serie...
example 2
Characterization of NLPs Produced By Cell-Free System
Solubilization of the bR-NLP Complex
[0133]The experimental design outlined in Example 1 of cell-free co-expression for refolding and incorporation into NLPs was also demonstrated using bR from Halobacterium salinarium, and truncated apolipoprotein A-1 (A 1-49) or Δ49A1. The bR protein is a seven transmembrane (TM) helical protein and serves as a structural model protein for rhodopsin and other 7-TM proteins such as GPCR family members.
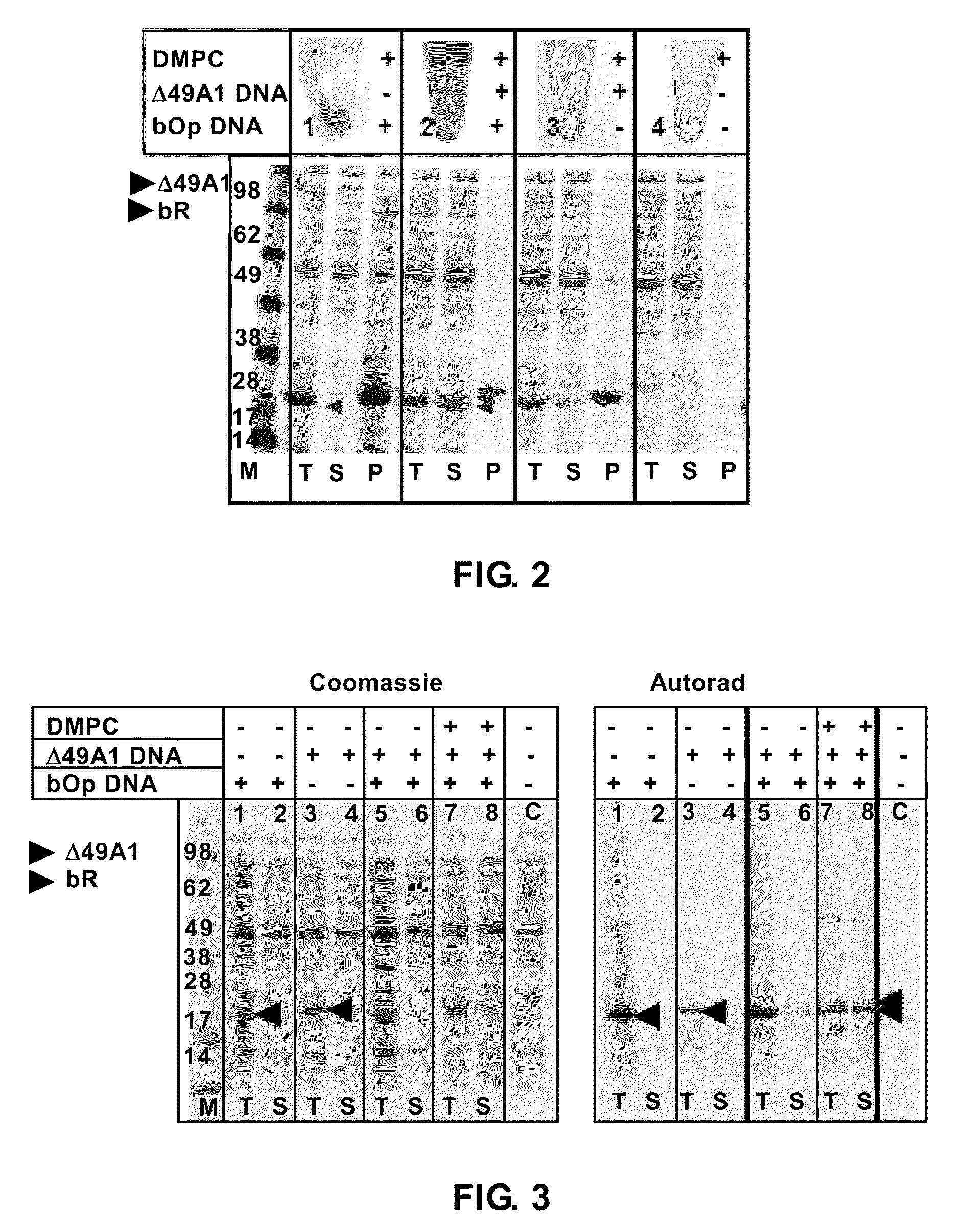
[0134]Simultaneous cell-free protein expression of both bR and Δ49A1 in the presence of DMPC in a single reaction produces a functional bR-NLP complex (FIG. 2 and FIG. 3).
[0135]In particular, as illustrated in FIG. 2, bOp & DMPC (sample 1) shows bR is insoluble in the absence of co-expression of Δ49A1; bOp, Δ49A1 co-expressed in the presence of DMPC (sample 2) shows bR remains in the soluble fraction with co-expressed Δ49A1; Δ49A1 & DMPC (sample 3) shows production of “empty”-NLPs; the control (sampl...
example 3
Characterization of NLPs produced by cell-free system
SDS Page,
Native Page SEC and AFM
[0139]bR-NLP complex heterogeneity was also observed by both native gel electrophoresis and SEC.
[0140]In particular, NLPs produced as described in Example 2 were first analyzed by SEC, to detect the separation of NLPs from larger lipid-rich material. Size exclusion chromatography identified a size shift in the bR-NLP complex compared to empty NLPs or liposomes. The bR-NLP complexes eluted primarily before empty NLPs and after liposomes (FIG. 5). A size range of approximately 470-680 kDa was observed for bR-NLP complexes, which was 160-370 kDa larger than the empty self-assembled NLPs (FIG. 6).
[0141]The NLPs were also analyzed by SDS Page. In particular, a 1 μL aliquot of the total (T) cell-free reaction, soluble (S) fraction and resuspended pellet (P) were diluted with 1×LDS Sample buffer with reducing agents (Invitrogen), heat denatured and loaded on to a 4-12% gradient pre-made Bis-Tris gel (Invit...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Electrical resistance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com