Methods, systems, and software for identifying functional biomolecules
a biomolecule and functional technology, applied in the field of molecular biology, molecular evolution, bioinformatics, etc., can solve the problems of inability to explore exhaustively, large protein sequence space, and long time-consuming to achieve protein design
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
character string variants that include multiple improved objectives relative to other members of the set of biopolymer character string variants.
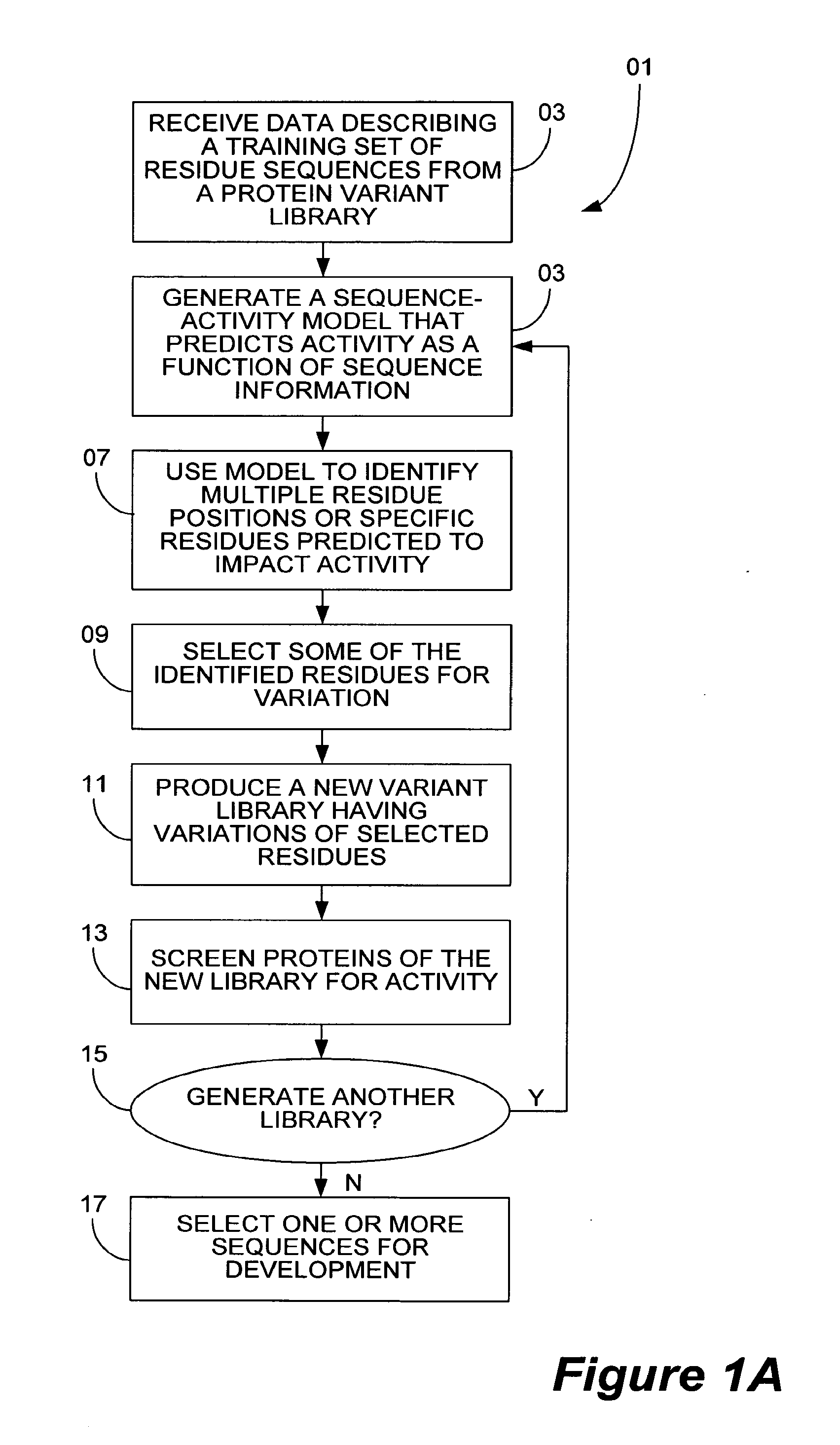
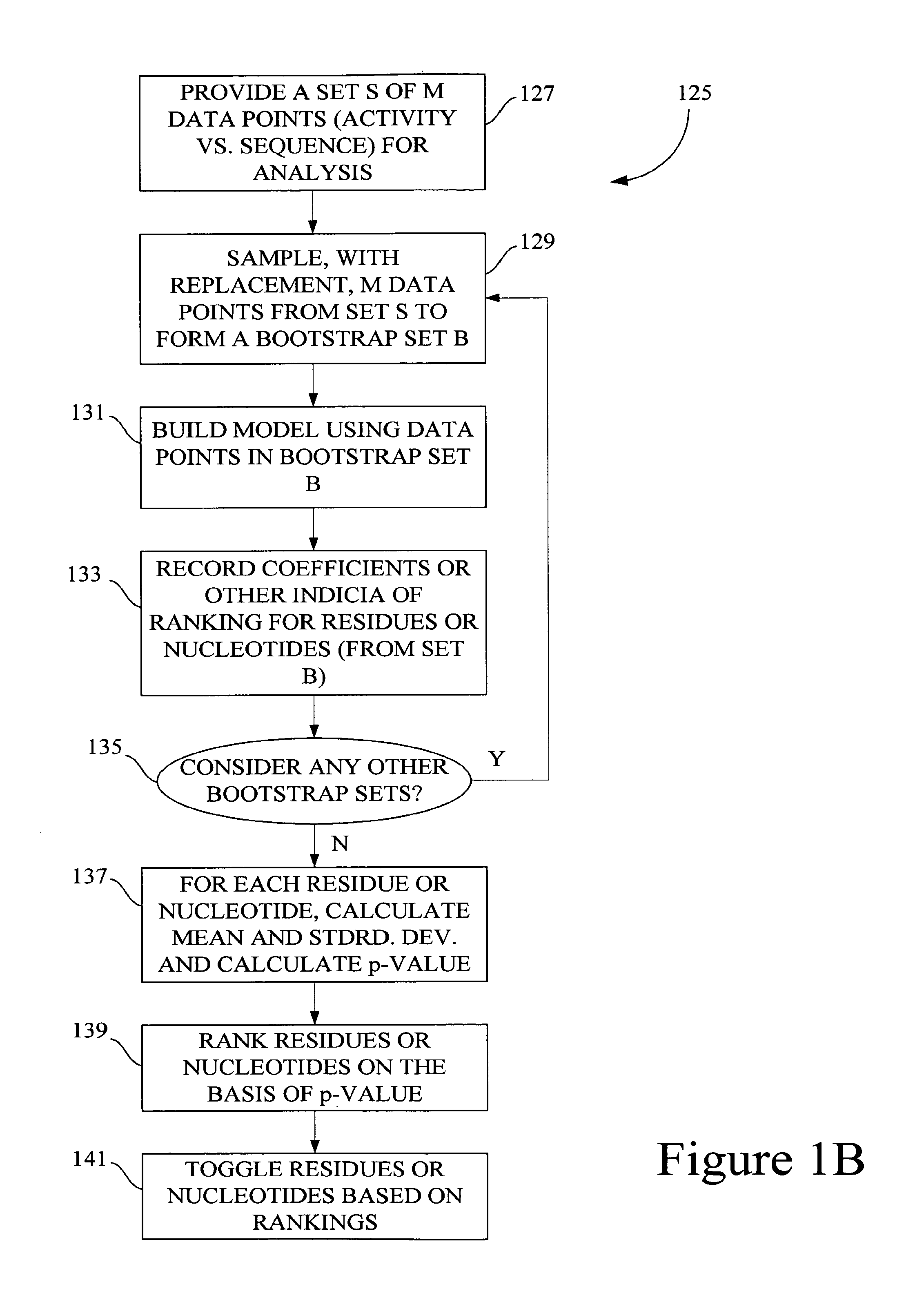
[0022]FIG. 6 is a chart that depicts steps performed in one embodiment of a method of evolving libraries for directed evolution.
[0023]FIG. 7 is a chart that depicts certain steps performed in an embodiment of a method of producing a fitter population of character string libraries.
[0024]FIG. 8 is a chart that shows certain steps performed in an embodiment of a method of selecting amino acid positions in a polypeptide variant to artificially evolve.
[0025]FIG. 9 is a chart that shows certain steps performed in another embodiment of a method of selecting amino acid positions in a polypeptide variant to artificially evolve.
[0026]FIG. 10 is a chart that shows certain steps performed in an embodiment of a method of identifying amino acids in polypeptides that are important for a polypeptide sequence-activity relationship.
[0027]FIG. 11 is a chart t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| physical | aaaaa | aaaaa |
| chemical properties | aaaaa | aaaaa |
| dimension | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com