Synthesis and enrichment of nucleic acid sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
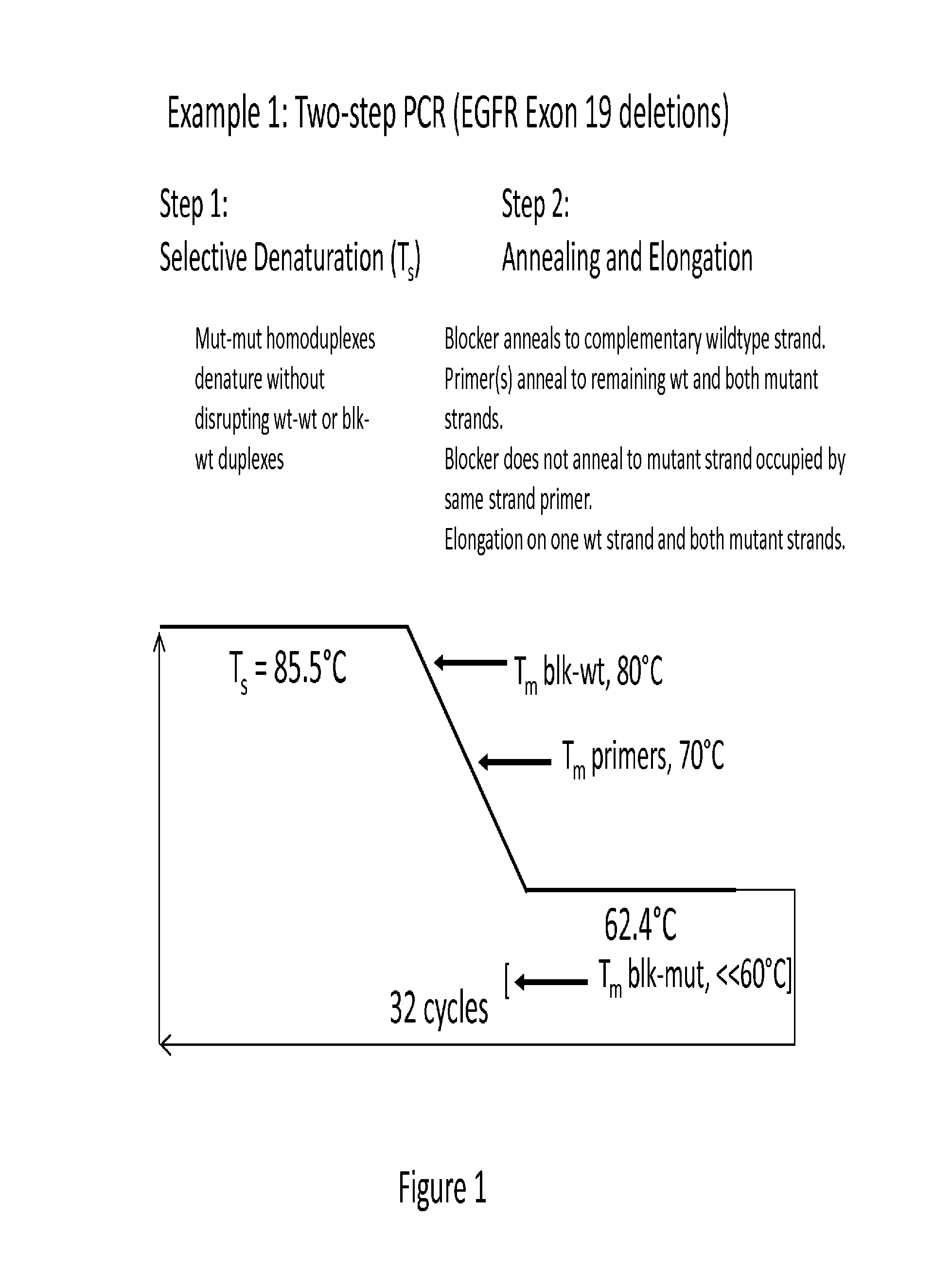
[0125]An example of a two-step PCR enrichment assay (EGFR Exon 19 deletions) is provided in FIG. 1. In this assay, a selective denaturation step precedes the annealing step. As the reaction ramps to the annealing temperature of 62.4° C., any complementary wildtype strands generated in the previous PCR cycle bind blocker before the primers anneal. The primers then anneal to the complementary mutant strand and bar the possibility of any blocker binding to that mutant strand due to sequence overlap. (For most deletions the likelihood of any blocker binding to the mutant is extremely low as there is actually very little common sequence). In this exemplary assay, at the preselected annealing step (62.4° C.), blocker:target and reference (wt):target heteroduplex formation is likely to be extremely low (below detectable or calculable levels) because only a minor percentage will be complementary.
TABLE 1Deletion-Specific Cycling Conditons (DSCC-3)StepTempTimeLid 98° C. Singe 1 (32 cycles)85...
example 2
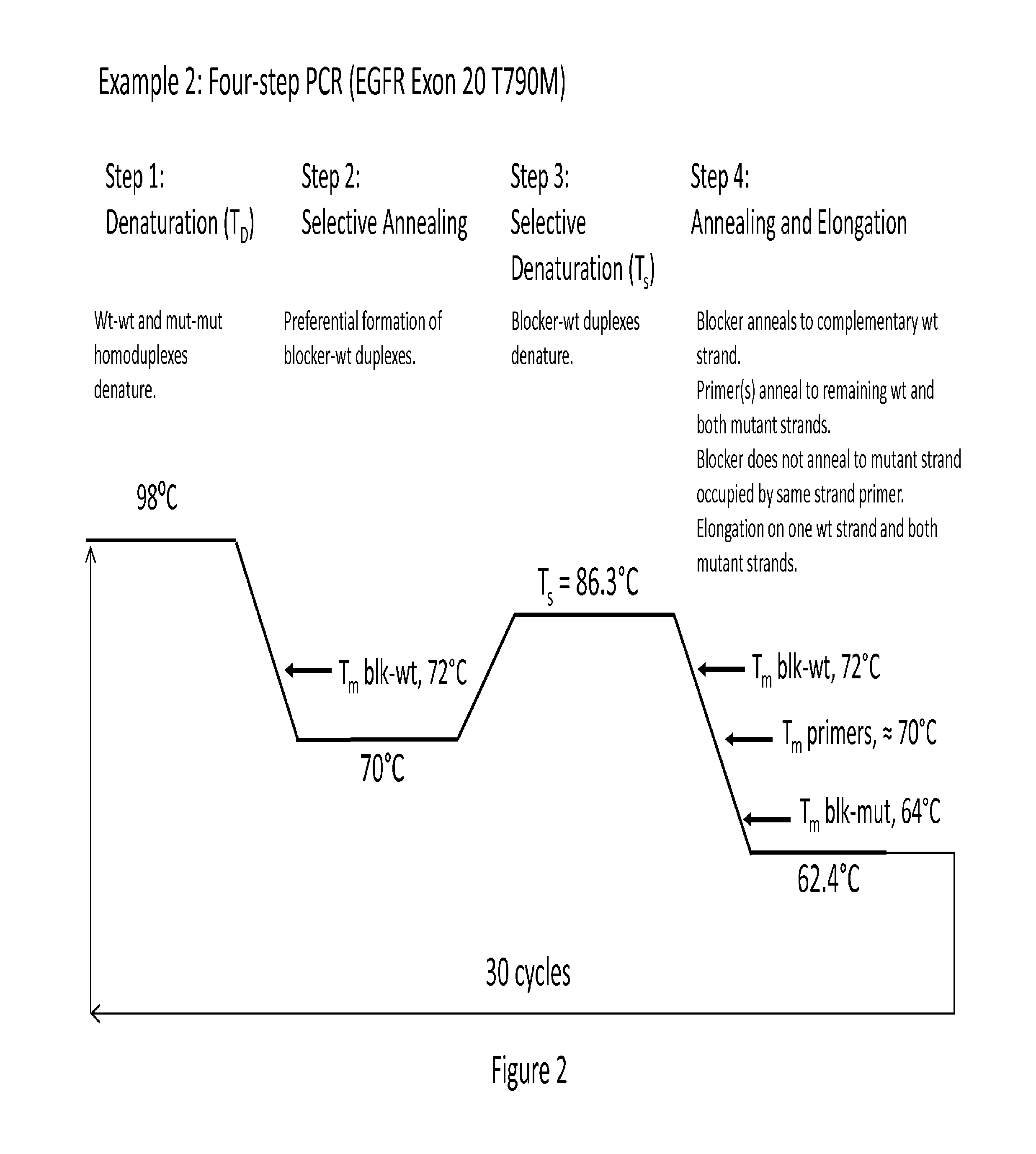
[0126]A schematic example of a four-step PCR enrichment assay (EGFR Exon 20 T790M) is provided in FIG. 2. In this assay, a 98° C. denaturation step ensures that all duplexes denature. During the second (optional) step (70° C.) blocker-wildtype duplexes form, but few blocker-mutant duplexes form (as 70° C. is above the blocker-mutant Tm). At step 3, selective denaturation, many of the blocker-wildtype will denature (along with any blocker-mutant duplexes that may exist). Just as in the two-step PCR (example 1), as the reaction ramps back to the annealing temperature of 64.0° C., complementary wildtype strands are bound by the excess blocker before the primers anneal. The primers then anneal to the complementary mutant strand and bar the possibility of blocker binding to that mutant strand. Short amplicon length allows extension without the need for an additional elongation step.
The level of enrichment for EGFR T790M_T is provided below in Table 6
TABLE 6FOLDENRICHMENTInputEGFR T790M_T...
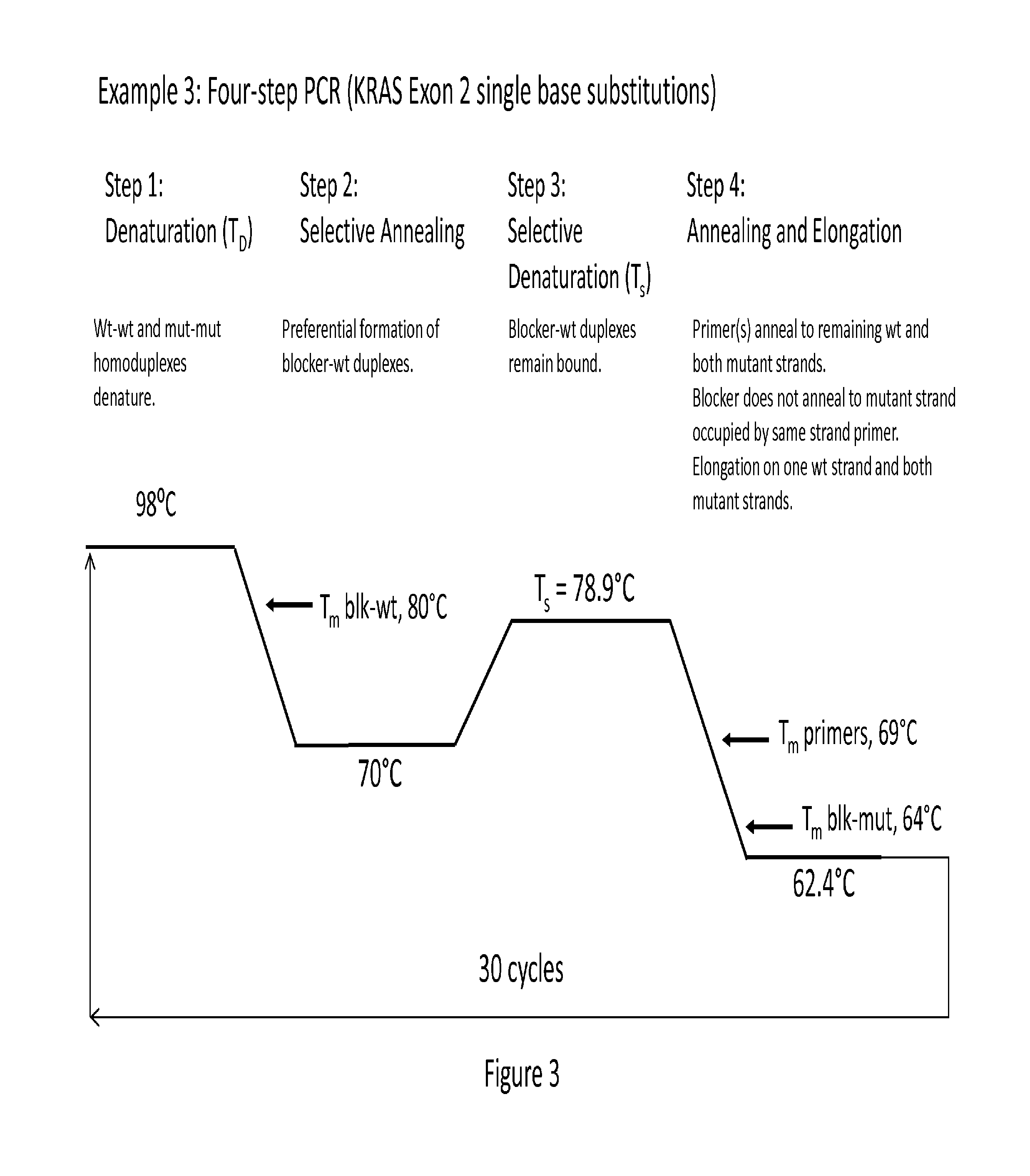
example 3
Verification of a Single Copy Assay Sensitivity
[0137]FIG. 6 provides a Normal distribution histogram and Poisson probabilities table. The table on the right is a Poisson distribution table of probabilities. The columns of the table are the number of observed events in a discrete interval. The rows are the average number of events per interval (0.13 events per interval, 0.25 events per interval, etc.) Each cell in the table is the probability of observing a given number of events (columns) given an average expected number of events (rows) in a single interval. For example, with regards to a cancer mutation detection test, if we expect to detect 2 mutants DNA strands per milliliter (interval), a single milliliter would have a 14% chance of containing no mutant DNA strands at all.
[0138]FIG. 7 Curve fit and calculated input mutation level of a cancer patient was detected in a biological fluid sample from the patient containing the KRAS G12D mutation. The raw data plot of the enriched re...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap