Characterizing prostate cancer
A technology for prostate cancer and prostate, applied in the field of methylation gene inspection, can solve problems such as small use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
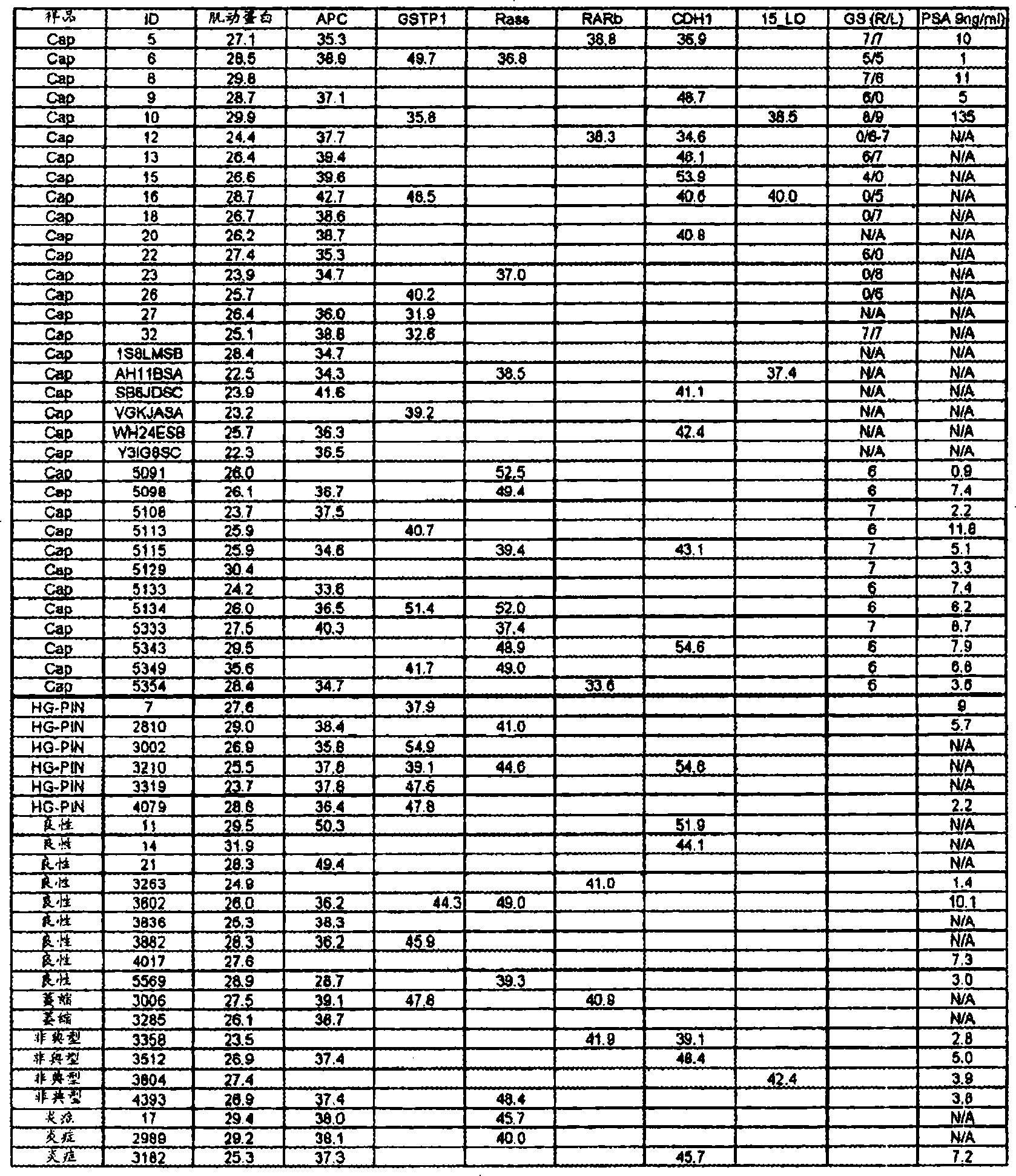
[0163] Example 1: Methylation Test and Gleason Score
[0164]Prostate samples were obtained from patients with known clinical outcomes. The Gleason score of the samples was determined according to known methods. According to these samples, the Gleason score of 52 samples was less than 7, the Gleason score of 36 samples was 7, and the Gleason score of 12 samples was greater than 7.
[0165] Methylation analysis was performed on each group using GSTP1 (Seq. ID No. 19, 20) and APC (Seq. ID No. 34, 35).
[0166] Methylation analysis was performed as follows. Genomic DNA was modified using a commercial sodium bisulfite transformation kit (Zymo Research, Orange, CA, USA). This treatment converts all cytosines in unmethylated DNA to uracil, while in methylated DNA only cytosines that are not in front of guanine are converted to uracil. All cytosines that precede guanines (in CpG dinucleotides) are still cytosines.
[0167] Sodium bisulfite-modified genomic DNA (100ng~150ng) was ...
Embodiment 2
[0177] Example 2: Serum Analysis
[0178] Serum samples were obtained from patients with known prostate cancer findings and from patients from whom biopsies were taken and Gleason scores were cited. Among these samples, 55 samples were from patients without cancer, 36 samples were from patients with Gleason score 5-6, and 21 samples were from patients with Gleason score 7-8.
[0179] The methylation status was determined according to the method in Example 1.
[0180] GSTP1 Marker was able to correctly detect methylation in 26% of samples from patients with Gleason score 7-8, and did not detect methylation in patients with Gleason score 5-6 or patients without cancer. Basicization. APC Marker was able to correctly detect methylation in 26% of samples from patients with Gleason score 7-8, and in up to 9 cases it also detected patients with Gleason score 5-6 or without cancer methylation in patients. The combined specificity of the two markers was 84%, the sensitivity was 18%...
Embodiment 3
[0206] Example 3: Urinalysis
[0207] Urine samples were obtained from patients with known prostate cancer findings and from patients from whom biopsies were taken and Gleason scores were cited. Among these samples, 42 samples were from patients with Gleason score 4-6 and 10 samples were from patients with Gleason score 7-9.
[0208] The methylation status was determined according to the method in Example 1 using Cepheid's Smart Cycler PCR equipment.
[0209] The combined specificity of the two markers, GSTP1 and RARb2, was 89% for post-massage urine samples and 91% for post-biopsy samples. The sensitivity of the methylation assay for post-massage samples was 40% in samples with Gleason scores below 7 and 78% in those samples with scores above 7. Thus, non-invasive sampling can be used in conjunction with other aspects of the invention.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com