Improved strategies for sequencing complex genomes using high throughput sequencing technologies
A genome sequence and genome technology, applied in the fields of molecular biology and genetics, can solve problems such as huge computing functions, time-consuming, expensive sequencing methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
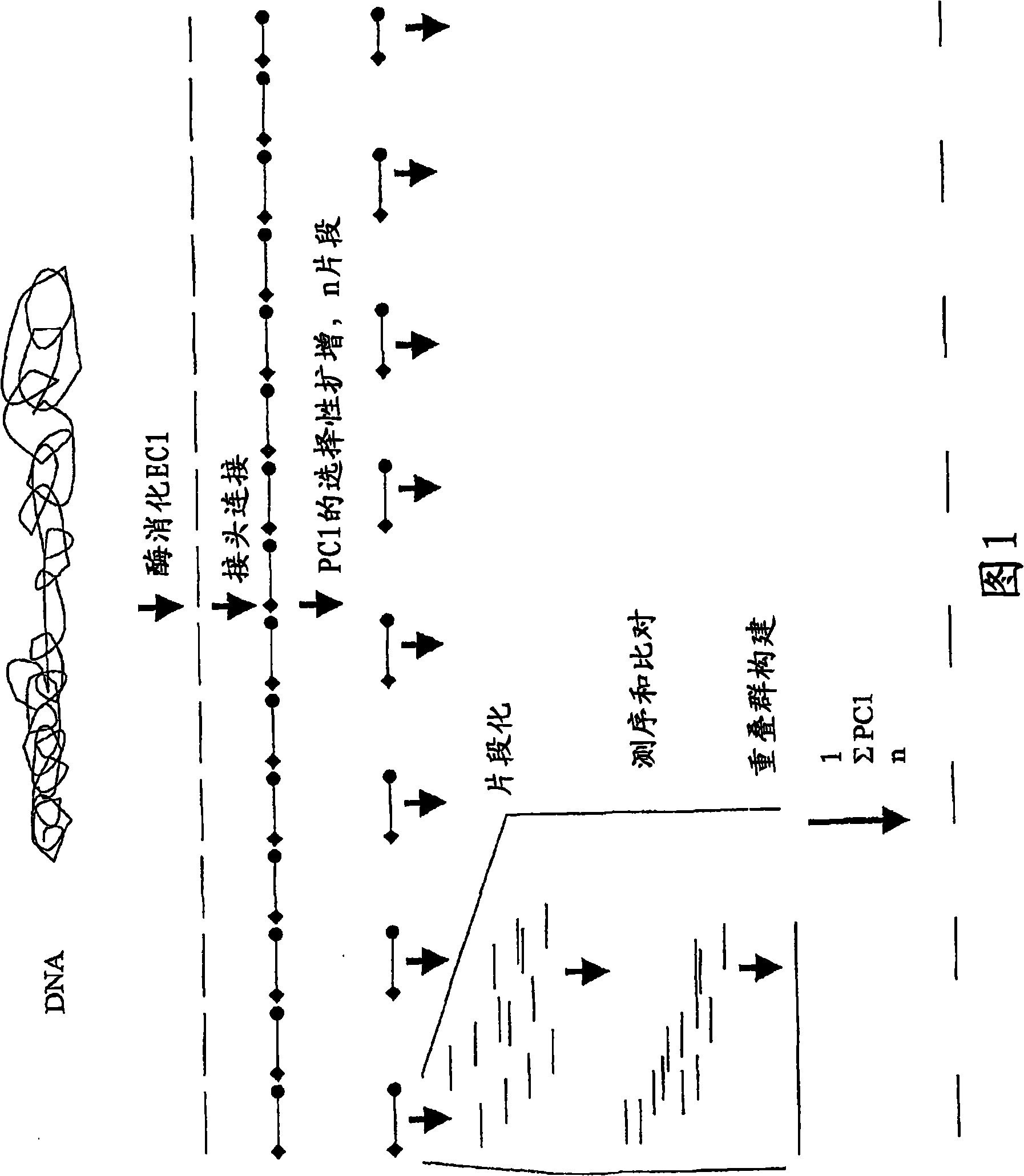
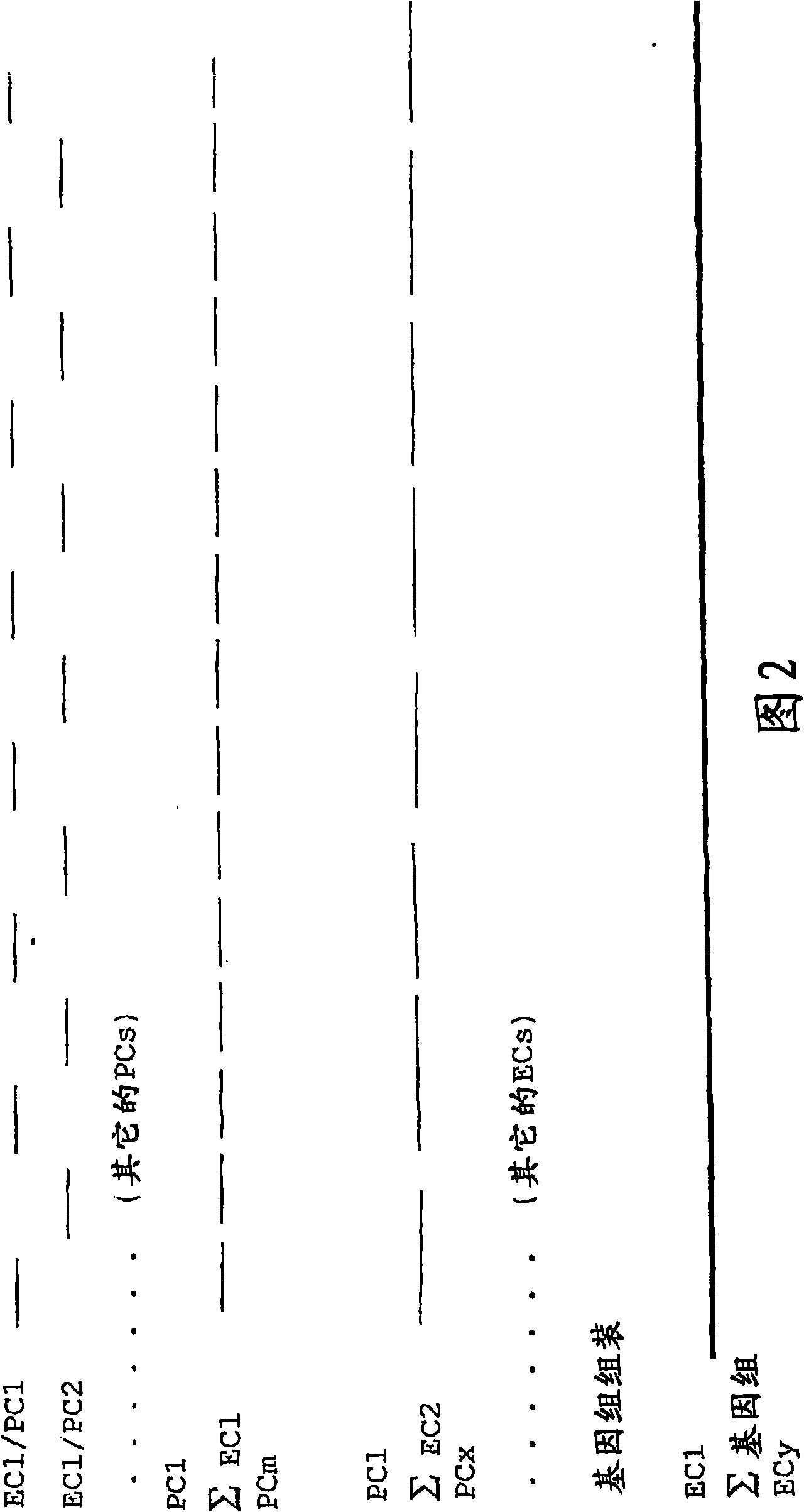
[0024] The invention provides a method for detecting genome sequence, which comprises the following steps:
[0025] (a) providing restriction fragments by digesting the genome with at least one first restriction endonuclease to provide a first subset of the genome;
[0026] (b) ligating at least one adapter to the first subset of restriction fragments to provide the first subset of adapter-ligated restriction fragments;
[0027] (c) selectively amplifying a first subset of adapter-ligated restriction fragments using a first primer combination to provide an amplified first subset of adapter-ligated restriction fragments, wherein at least the first primer comprises a primer complementary to the adapter and A fragment complementary to a part of the restriction endonuclease recognition sequence, further comprising a first selected sequence at the 3' end of the primer sequence, wherein the first selected sequence comprises 1-10 selected nucleotides;
[0028](d) repeating step (c) ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com