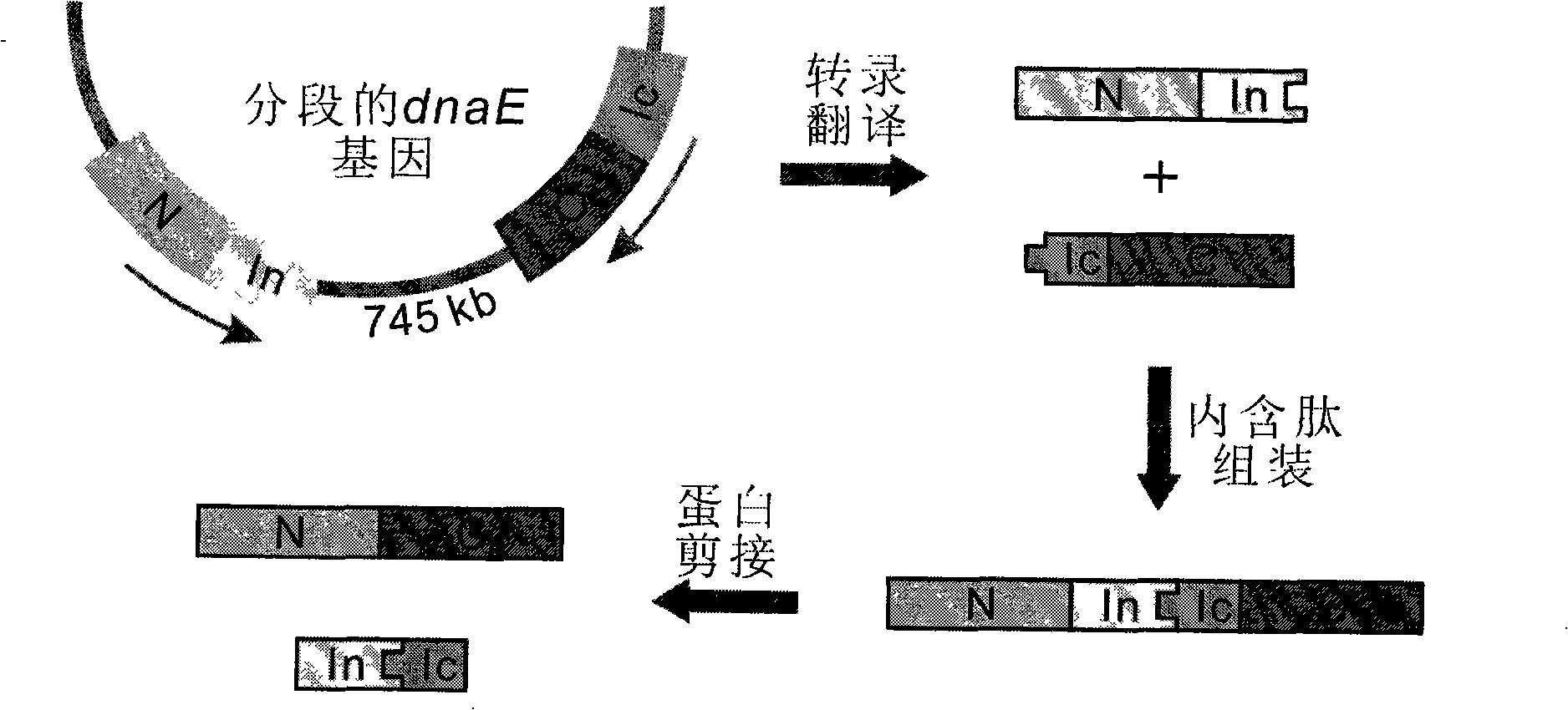
Method using implicit peptides trans splicing to manufacture protein molecular weight standard
A molecular weight standard, protein technology, applied in the field of biological science and biology, can solve the problem of high production cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
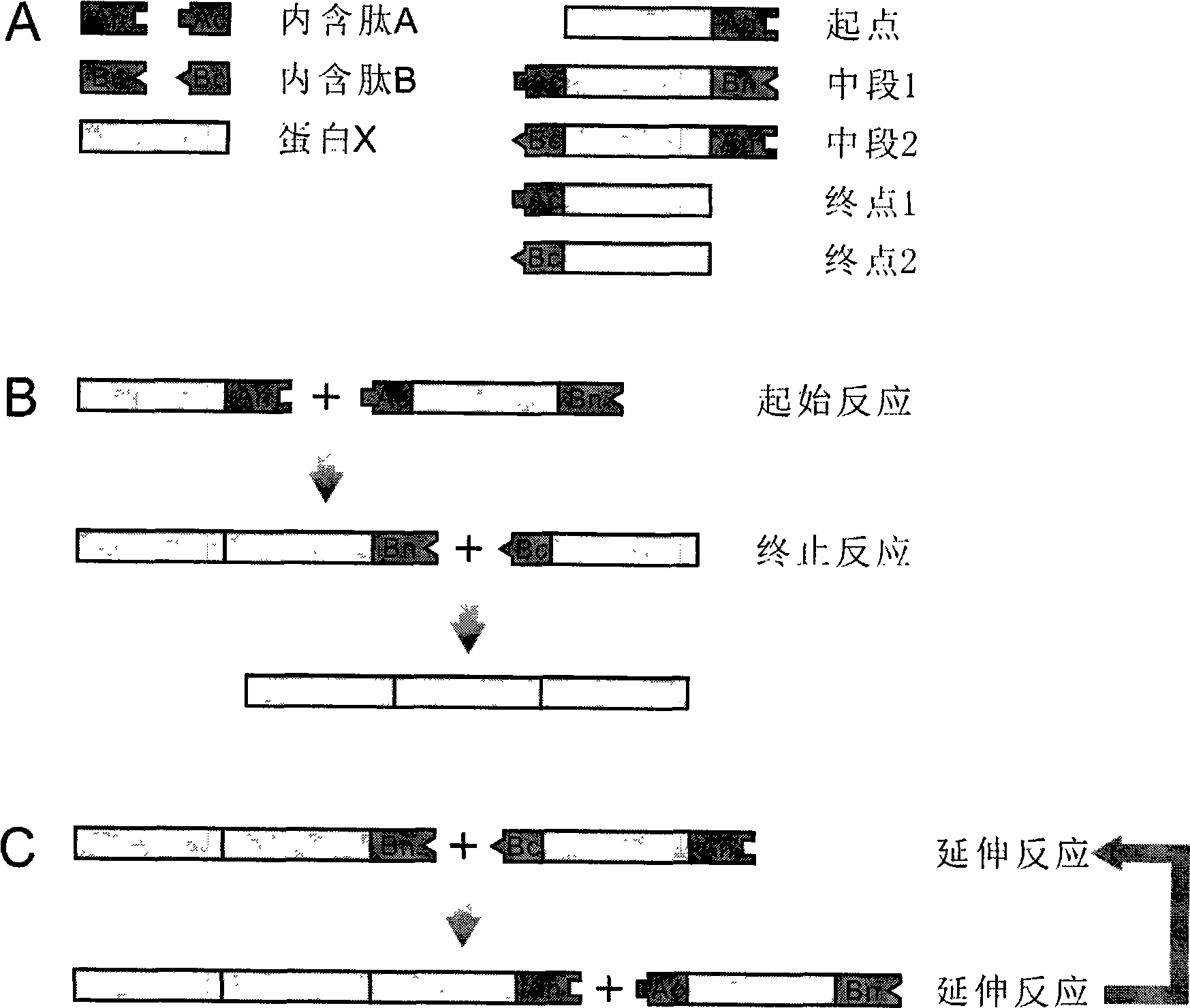
[0018] For example, DnaE and DnaB of the cyanobacteria Synechocystis sp. PCC6803 decompose intein A and B into amino terminal (N terminal) and carboxyl terminal (C terminal), namely An, Ac, Bn, Bc. They are then combined into five recombinant proteins, starting point (X-An), middle segment 1 (Ac-X-Bn), middle segment 2 (Bc-X-An), end point 1 (Ac-X), end point 2 (Bc- X).
[0019] First let the "starting point" protein react with the "end point 2" protein to obtain a protein with twice the molecular weight of the known protein;
[0020] If the "starting point" protein and the "middle segment 2" protein are reacted, and then the "end point 1" protein is added to the reaction, a protein with a molecular weight three times that of the known protein can be obtained;
[0021] If the "starting point" protein and the "middle segment 1" protein are reacted, then the "middle segment 2" protein is added for reaction, and the "end point 2" protein is finally added, a protein with a molecu...
PUM
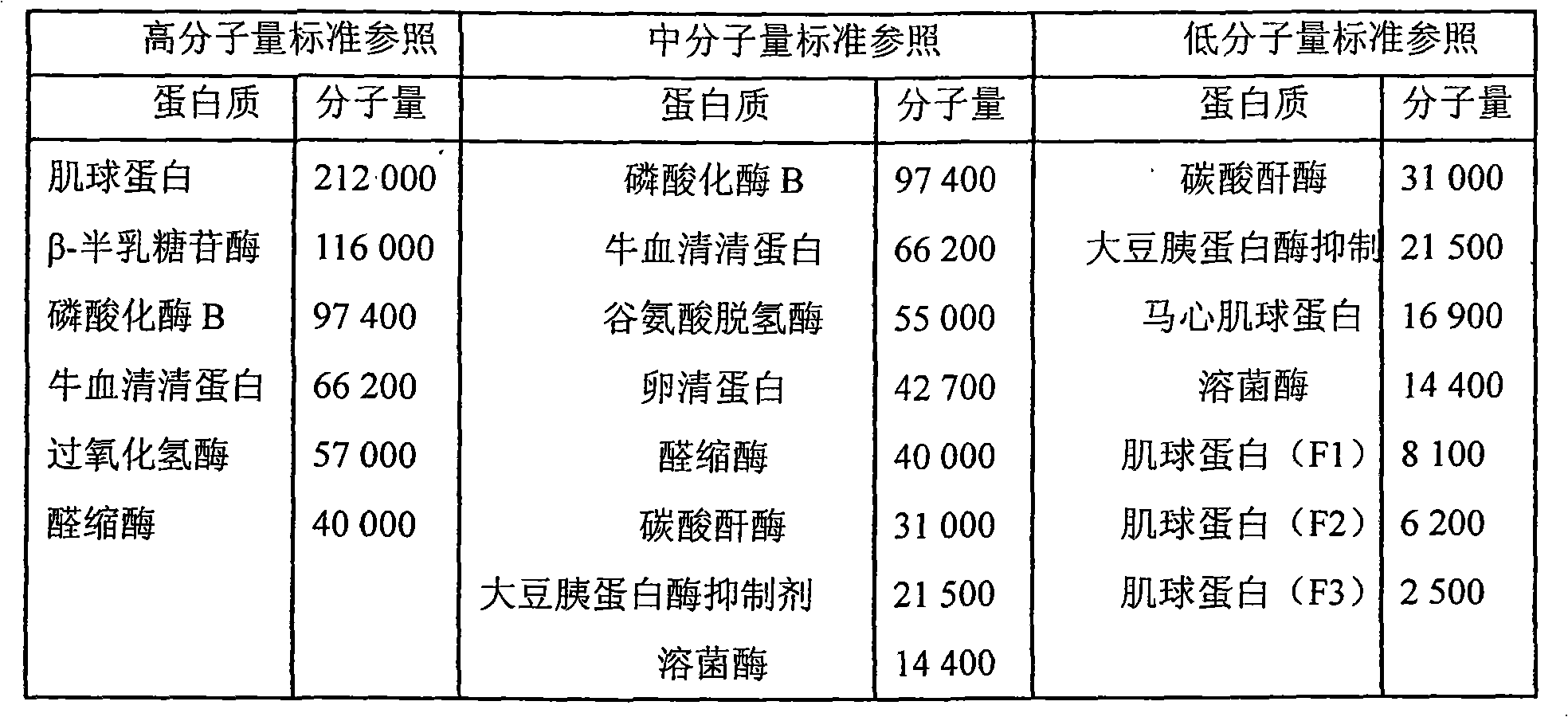
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com