Cyanobacteria modified by gene engineering and use thereof for producing ethanol
A technology of cyanobacteria and base sequence, which is applied in the field of genetically engineered cyanobacteria and its production of ethanol, and can solve the problems of insufficient concentration of ethanol production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
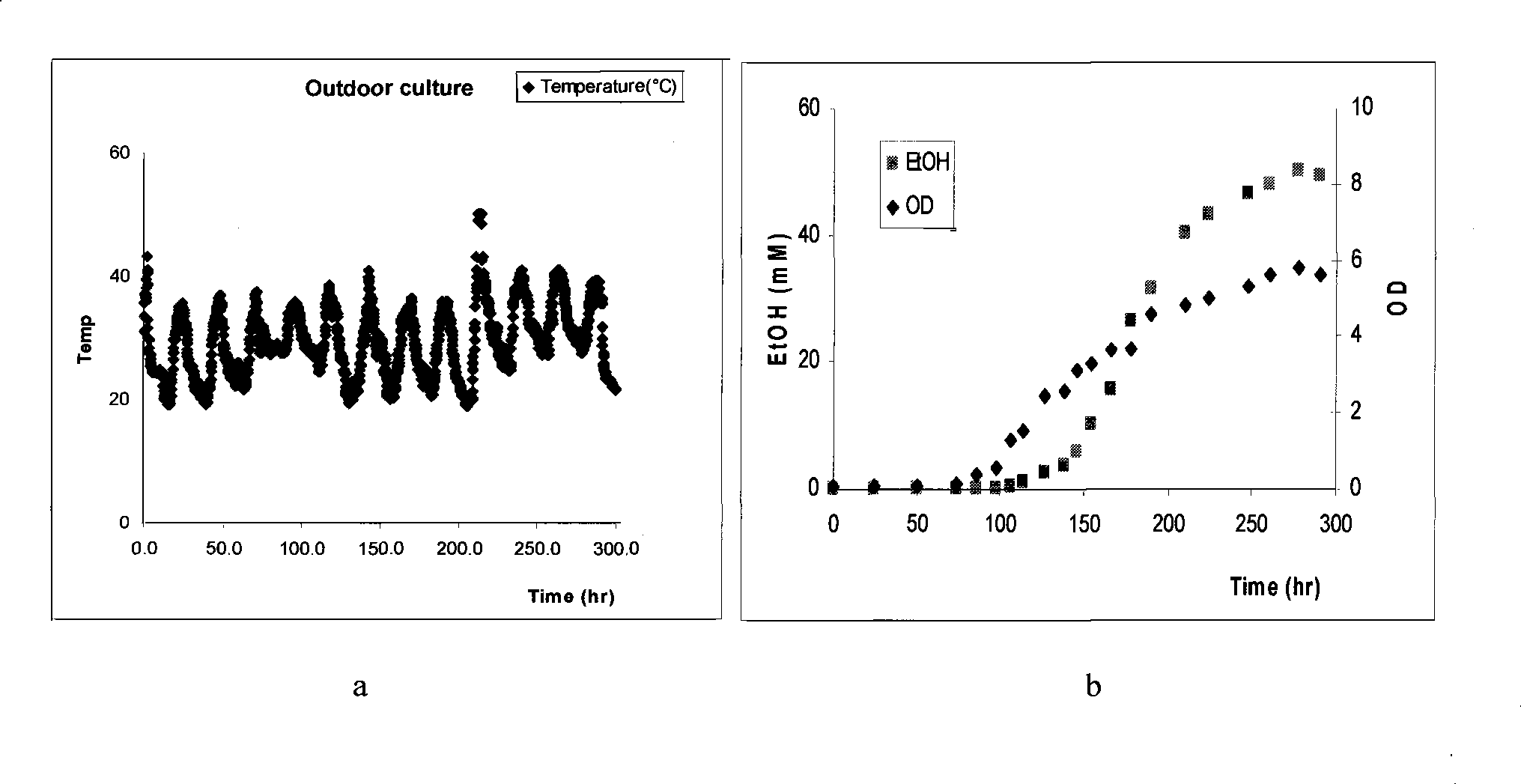
[0050] Specific operation: the wild Synechocystis sp. 6803 was inoculated in the photosynthetic bioreactor group (consisting of six 150ML shake flasks built in a modified shaker, and the working solution was 50ML), and it was cultured by BG-11 culture medium, and the shaker had a built-in light source , the light intensity on the surface of the reactor is about 200Einstein / m 2 / s. The temperature of the shaker was regulated below 30 °C and the speed was 200 RPM. After 5 days, put them into a hot water bath box to heat and heat shock. Then put it back into the photosynthetic bioreactor at 30°C, and after the complete "whitening" phenomenon occurs (about 3 days), centrifuge the Synechocystis 6803 cells, discard the supernatant, and use fresh BG-11 culture medium to cultivate " albino" of Synechocystis 6803 cells. Synechocystis cells adapted to the heat shock will regenerate chlorophyll, thereby restoring photosynthesis and growth. At the same time, the metabolic mechanism of...
Embodiment 2
[0051] Embodiment 2: pyruvate decarboxylase gene pdc and nirA promoter design
[0052] By the method of bioinformatics, be easy to obtain the sequence of pyruvate decarboxylase gene pdc and nirA promoter (the inventor of the present invention uses the search engine http: / / www.kegg.com / that Japanese Kyoto Encyclopedia of Genes and Genomes provides The sequences of the pyruvate decarboxylase gene pdc and the nirA promoter were obtained). Pdc (sequence number: 1) and nirA (sequence number: 2) can be obtained by artificial synthesis in turn (Roy, et al. European Journal of Biochemistry.191 (3): 647-652, 1990; Shabalina, et al. Nucleic Acids Res. 34:2428-2437, 2006; Satya et al. Proc IEEE Comput Soc Bioinform Conf. 2:294-305, 2003; Kim, et al. Gene. 199:293-301, 1997). The sequence obtained above was analyzed by the Sanger method (F. Sanger, Science, 214, 1215, 1981) to confirm the base sequence.
Embodiment 3
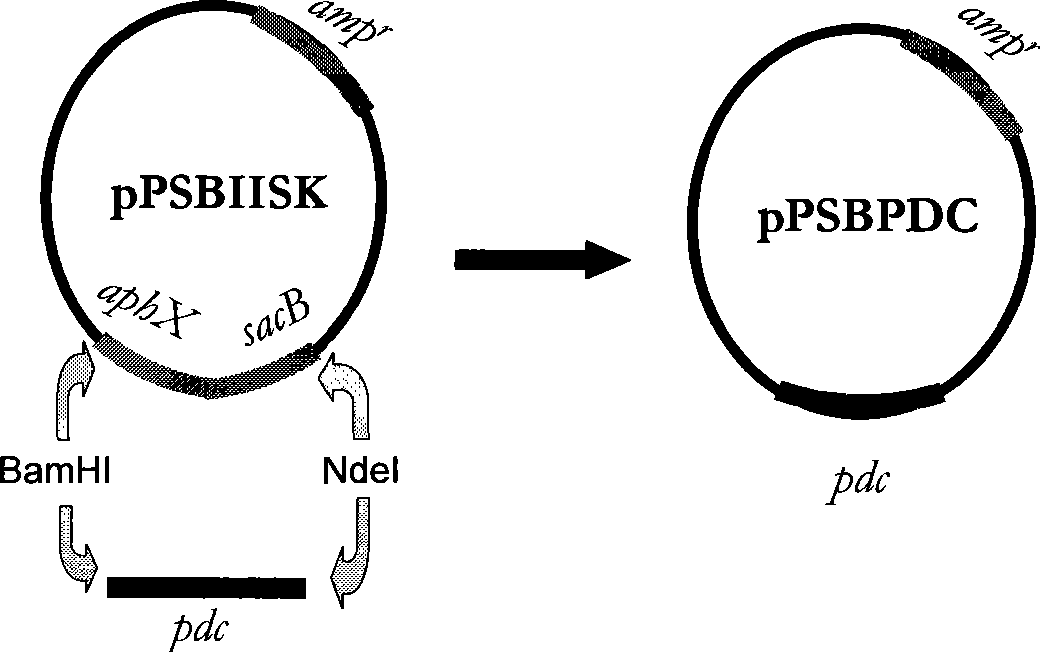
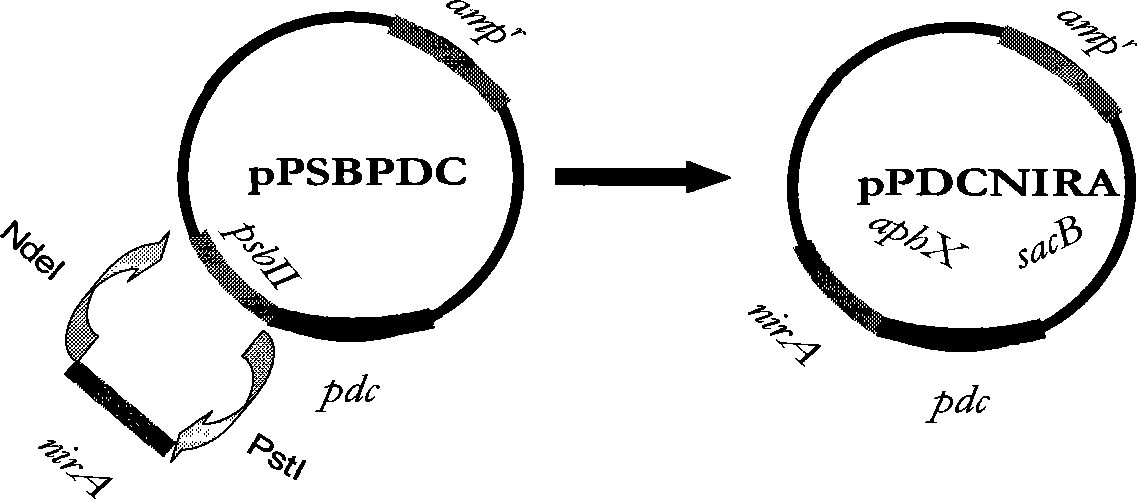
[0053] Embodiment 3: Transformation of Synechocystis mutant strain S.strictus with pdc gene
[0054] A pair of PCR primers P1 (sequence number: 4) and P2 (serial number: 5):
[0055] P1 (5-terminal primer): 5'-GGAGTAAG CATATG AGTTATACTG-3'
[0056] Nd
[0057] P2 (3' end primer): 5'-GGATCTCGACTCTAGA GGATCC -3'
[0058]BamHI
[0059] Among them, a NdeI restriction site was designed for primer P1, and its sequence was CA↓TATG, and a BamHI restriction site was designed for primer P2, and its sequence was GGATC↓C. A pdc DNA fragment P of 1410 bp was amplified by conventional PCR amplification method. PCR reaction: add in 0.5mL sterile centrifuge tube successively: deionized water 36μl; 10XTaq enzyme buffer 5μl; 4XdNTP (2.5mmol / L) 5μl; P1 primer 1μl; P2 primer 1μl; ) 1 μl; Taq enzyme (3U / μl) 1 μl. A total of 50 μl.
[0060] Thermal cycle parameters of PCR reaction: first, 94°C pre-denaturation for 5 minutes; then 94°C denaturation for 30 se...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com