Method for detecting DNA fingerprint purity in cotton crossbreeds
A technology of purity detection and hybridization, which is applied in the field of molecular biology, can solve the problems of lack of systematic research on variety identification, insufficient identification ability, and high experimental cost, and achieve stable and reliable detection results, which is conducive to standardization and simple and convenient operation. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
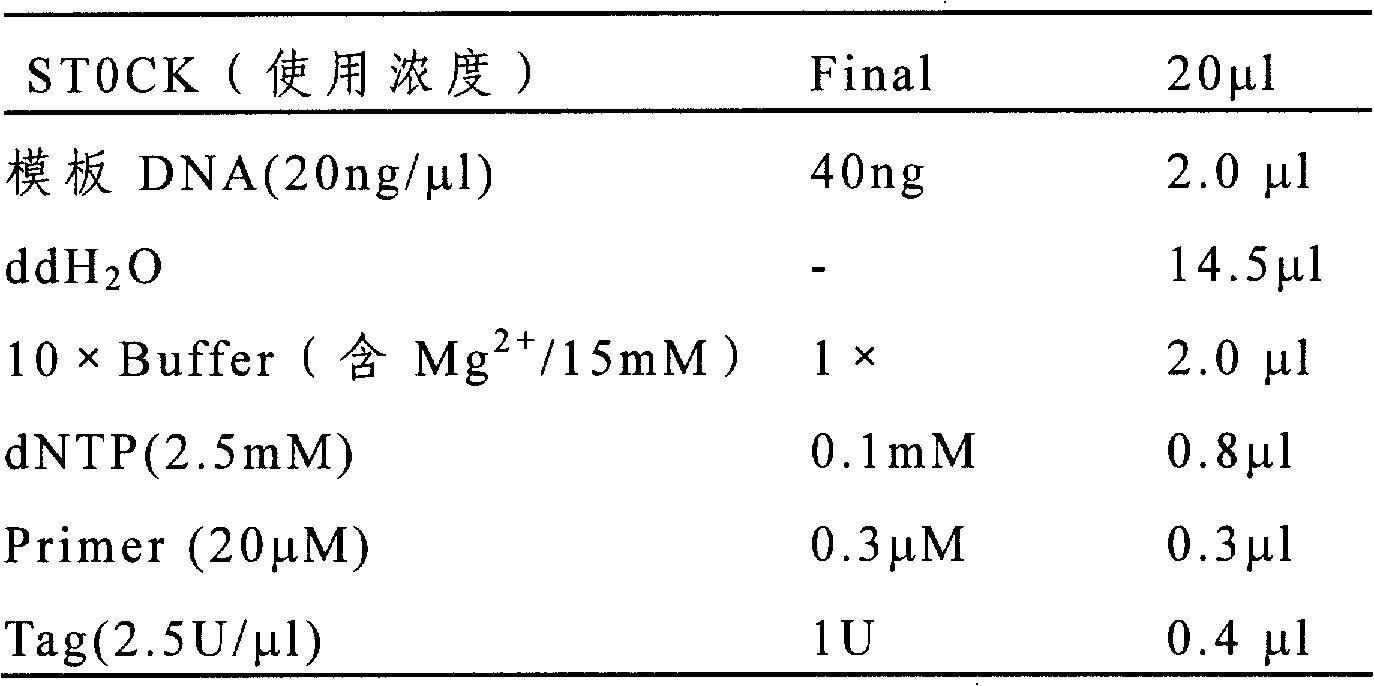
[0030] The purity detection of the 48 seed samples of China Cotton Research Institute includes the following steps. Among them, the primer information used in this method is shown in Table 1; the reagent preparation used is shown in Table 2.
[0031] 1. Single grain cotton DNA extraction
[0032] (1) Shell a single cotton seed, thoroughly pulverize it and transfer it to a 2ml centrifuge tube.
[0033] (2) Add 800ul DNA extraction solution (1% SDS, 0.01M EDTA 8.0, 0.7M NaCl, 0.05M Tris-HCl, 0.5% sorbitol, 1% PVP, 1% β-mercaptoethanol), vortex until fully mixed , in a 65°C water bath for 30 minutes, and shake gently at intervals of about 10 minutes.
[0034] (3) After the water bath is over, add an equal volume of 800 μl of a mixture of phenol, chloroform and isoamyl alcohol (the volume ratio is 2-5:24:1 in turn), mix up and down until there is no layering, and centrifuge at 10,000 rpm for 10 min.
[0035](4) Transfer the supernatant to another 2ml centrifuge tube, add 1uL RNa...
Embodiment 2
[0069] According to the steps of Example 1, the purity of the 48 seed samples of China Cotton Research Institute was tested using the characteristic primer BNL1705. The results of data statistics and analysis are as follows:
[0070] figure 2 Shown is the purity test result of the characteristic primer BNL1705 on 20 samples of 48 seeds of China National Cotton Research Institute, and the control figure 1 The male parent, female parent and hybrid F of Zhongmian Institute 48 shown in 1 The standard electrophoresis bands of the standard electrophoresis bands show that: ①Paternal band type: sample No. 16; ②Maternal band type: sample No. 7 and No. 20; ③Other hybrid band types: sample No. 13; ④True hybrid band type: except the above Except for the banding patterns of the three hybrids, the other 16 samples were all banding patterns of true hybrids. Therefore, the purity of these 20 Zhongmian Institute 48 seed samples is 80%; the rate of hybrid plants is 20% (wherein the rate of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com