Anti-PCVD shRNA and design-synthesis method and application thereof
A technology of porcine circovirus and virus, applied in antiviral agents, DNA/RNA fragments, DNA preparation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
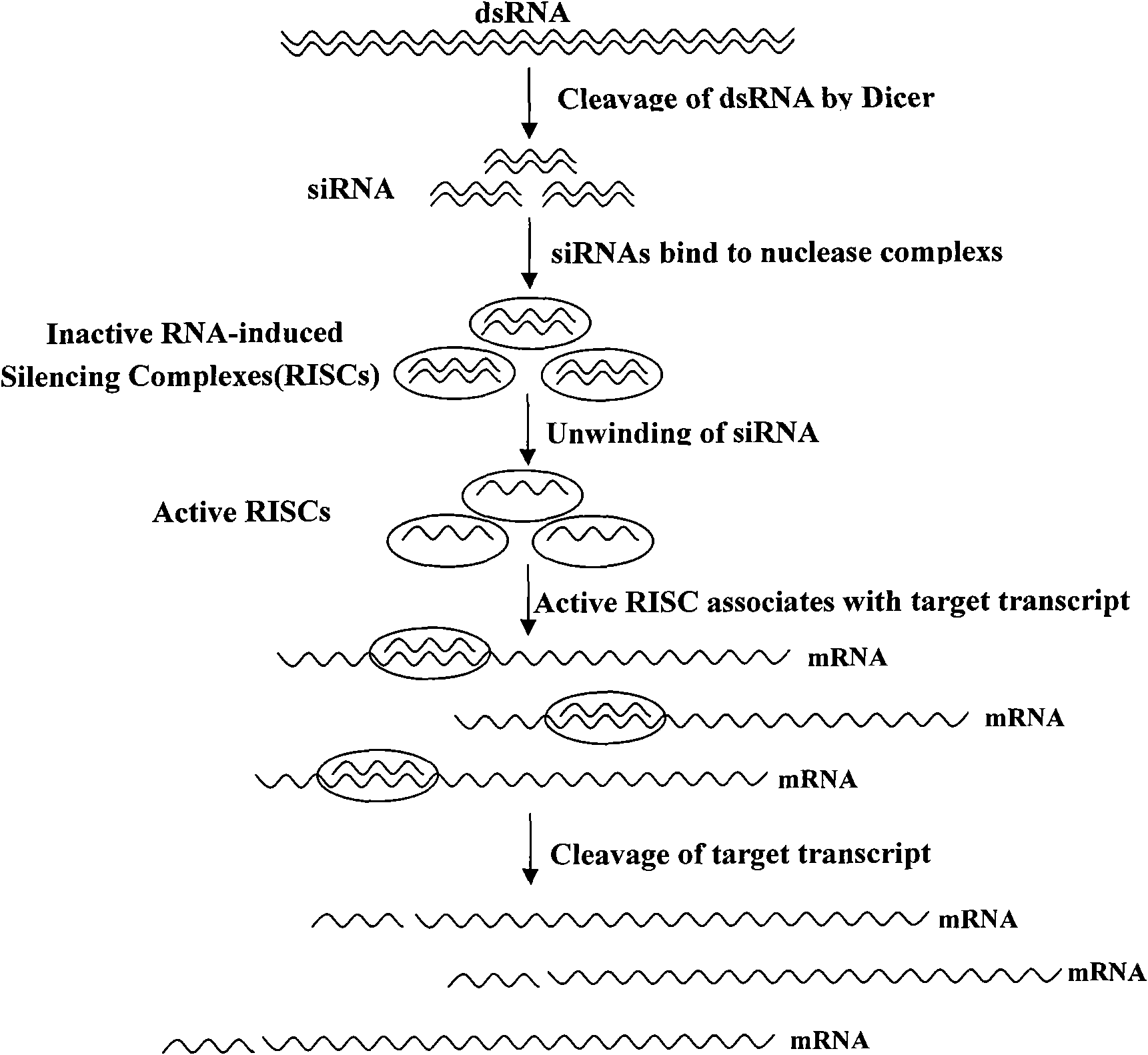
[0197] Embodiment 1, the selection of anti-porcine circovirus disease shRNA
[0198] 1. According to the genome sequence of PCV-2 type virus, take Rep gene and Cap gene as target gene, select a section of 19bp long nucleic acid sequence. The sequence should not be in the 5' and 3' untranslated regions, and 75 bp after the start codon, because these regions are rich in regulatory proteins, which may affect the effect of RNA interference.
[0199] 2. Calculate the GC content of the 19bp nucleic acid sequence, and the GC content must be between 40% and 60%. The GC content is optimal around 45%.
[0200] 3. There are at least 3 A or T nucleic acid residues in the 15-19 position, which can enhance the RNA interference activity.
[0201] 4. Check the 19bp nucleic acid sequence to ensure that it will not form a secondary structure.
[0202] 5. The 19bp nucleic acid sequence that simultaneously meets the three conditions of step 2, step 3 and step 4 is used as a candidate nucleic a...
Embodiment 2
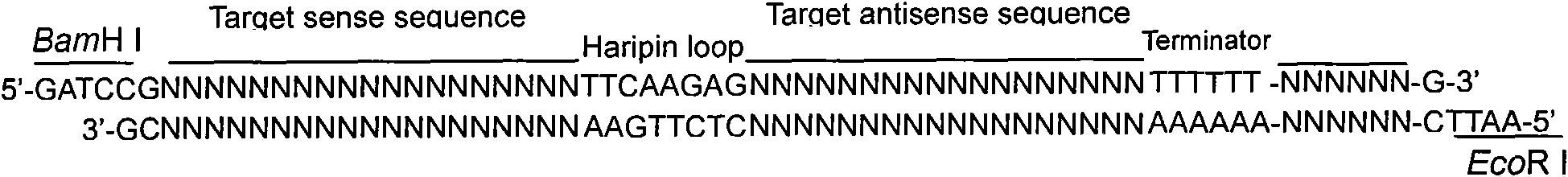
[0203] Embodiment 2, the design and synthesis of anti-porcine circovirus disease shRNA
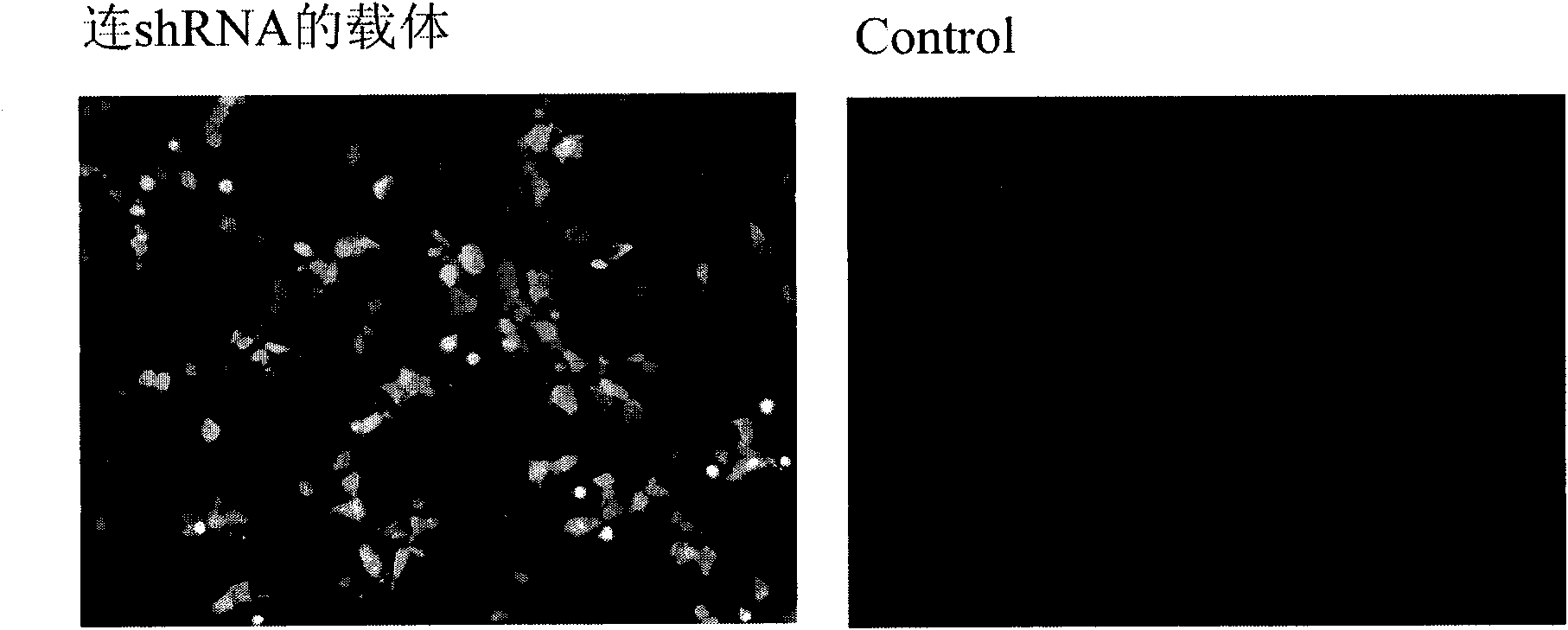
[0204] According to the RNA interference vector we used, RNAi-Ready pSIREN-RetroQ-ZsGreen Vector, the designed shRNA sequence was annealed (95°C / 30sec, 72°C / 2min, 37°C / 2min, 25°C / 2min) to form a sticky The double strand at the end can be directly ligated to the linearized RNAi-Ready pSIREN-RetroQ-ZsGreen vector (known vector).
Embodiment 3
[0205] Embodiment 3, the anti-porcine circovirus shRNA of design and synthesis
[0206] PSIREN-REP-1:
[0207] 5'-GATCCGTACGGGAGCTTCCAATCTCTTCAAGAGAGAGATTGGAAGCTCCCGTATTTTTTACGCGTG-3'
[0208] 3'-GCATGCCCTCGAAGGTTAGAGAAGTTTCTCTCTCTAACCTTCGAGGGCATAAAAAATGCGCACTTAA-5'
[0209] PSIREN-REP-2:
[0210] 5'-GATCCAGTGAGCGGGAAAATGCAGTTCAAGAGACTGCATTTTCCCGCTCACTTTTTTTACGCGTG-3'
[0211] 3'-GTCACTCGCCCTTTTACGTCAAGTTCTCTGACGTAAAAGGGCGAGTGAAAAAAAATGCGCACTTAA-5'
[0212] PSIREN-REP-3:
[0213] 5'-GATCCGTGGTACTCCTCAACTGCTGTTCAAGAGACAGCAGTTGAGGAGTACCATTTTTTACGCGTG-3'
[0214] 3'-GCACCATGAGGAGTTGACGACAAGTTCTCTGTCGTCAACTCCTCATGGTAAAAAATGCGCACTTAA-5'
[0215] PSIREN-REP-4:
[0216] 5'-GATCCAATGCAGAAGCGTGATTGGTTCAAGAGACCAATCACGCTTCTGCATTTTTTTACGCGTG-3'
[0217] 3'-GTTACGTCTTCGCACTAACCAAGTTCTCTGGTTAGTGCGAAGACGTAAAAAAAATGCGCACTTAA-5'
[0218] PSIREN-REP-5:
[0219] 5'-GATCCAAGCAAATGGGCTGCTAATTTCAAGAGAATTAGCAGCCCATTTGCTTTTTTTTACGCGTG-3'
[0220]3'-GTTCGTTTACCCGACGATTAAAGTTCTCTTAATCGTCGGGT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com