Methods of diagnosing rejection of a kidney allograft using genomic or proteomic expression profiling
A graft rejection, allogeneic technology, applied in the fields of biochemical equipment and methods, disease diagnosis, microbial determination/examination, etc., can solve the problem of not giving kidney transplant reviews and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0109]For the preparation of monoclonal antibodies against biomarkers, any of the techniques provided for the production of antibody molecules can be used. Such techniques include, but are not limited to, hybridoma technology, triple hybridomas (e.g. Kohler and Milstein 1975, Nature 256:495-497; Gustafsson et al., 1991, Hum. Antibodies Hybridomas 2:26-32), human B- Cell hybridomas or EBV hybridomas eg (Kozbor et al., 1983, Immunology Today 4:72; Cole et al., 1985, in: Monoclonal Antibodies and Cancer Therapy, Alan R. Liss, Inc., pp. 77-96). Human or humanized antibodies can be used and can be obtained by using human hybridomas (Cote et al., 1983, Proc. Natl. Acad. Sci. USA 80:2026-2030) or by in vitro transformation of human B cells with EBV virus (Cole et al., 1985, see: Monoclonal Antibodies and Cancer Therapy, Alan R. Liss, Inc., pp. 77-96). Techniques developed for the production of "chimeric antibodies" (Morrison et al., 1984, Proc. Natl. Acad. Sci. USA 81:6851-6855; Neu...
Embodiment approach
[0243] Nucleic acid profiling can also be used in combination with metabolite ("metabolomics") or proteomic profiling. Minor alterations in the subject's genome (eg, single nucleotide changes or polymorphisms) or genomic expression (eg, differential gene expression) may result in a rapid response in the subject's small molecule metabolite profile . Small-molecule metabolites can also respond rapidly to environmental changes, with significant metabolite changes occurring within seconds to minutes of environmental change—conversely, changes in protein or gene expression may take hours or days to manifest. The list of clinical variables includes, for example, cholesterol, homocysteine, glucose, uric acid, malondialdehyde, and ketone bodies. Additional non-limiting examples of small molecule metabolites are listed in Table 3.
[0244] Table 3: Metabolites identified and quantified in NMR spectra of serum samples obtained from subject populations
[0245] Compound name ...
Embodiment 1
[0287] Example 1: Comparison of Biomarkers and Clinical Diagnosis
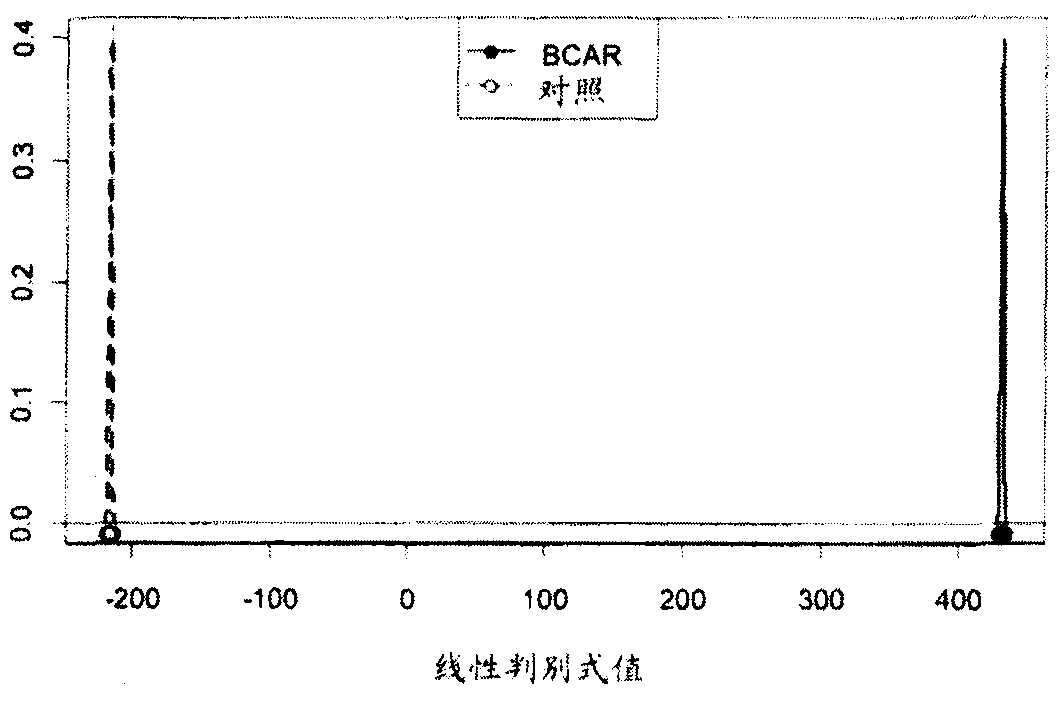
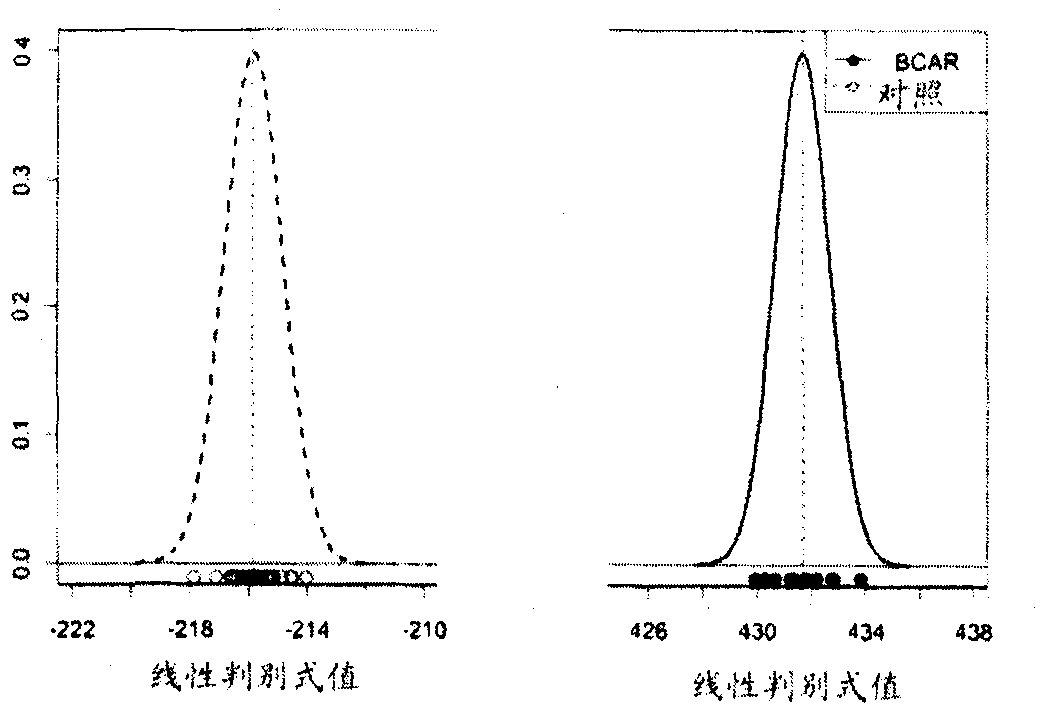
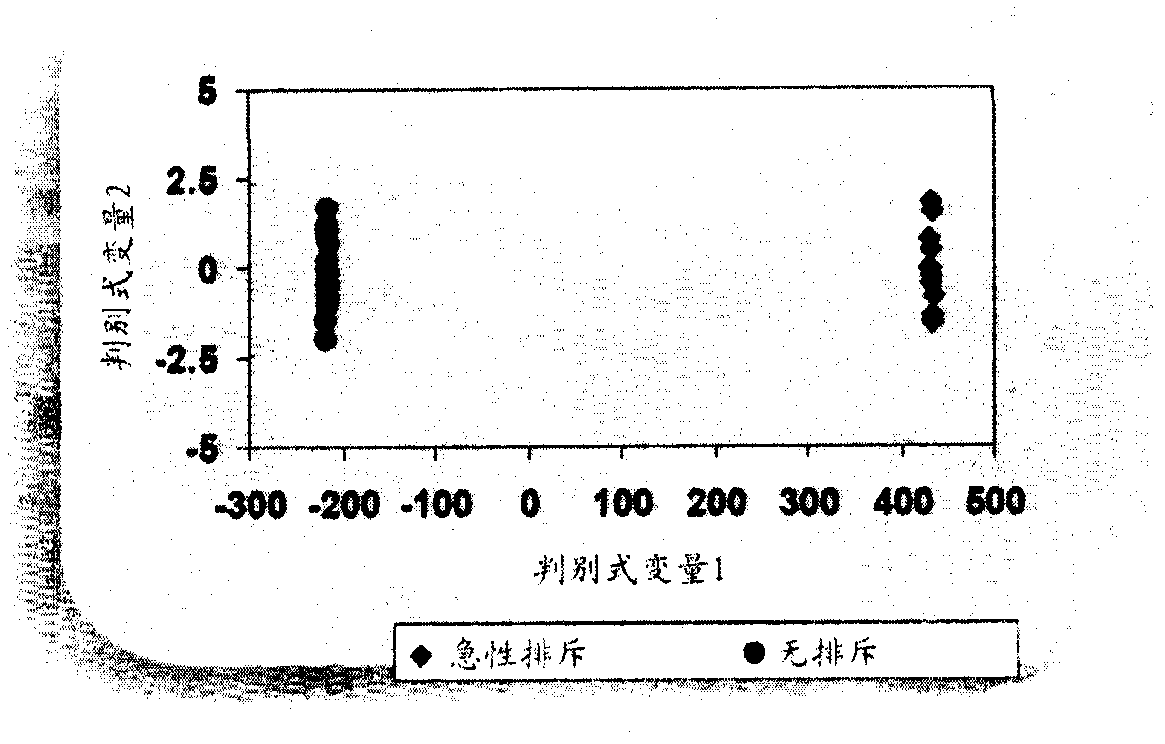
[0288] A total of 33 subjects were included in the study, including 11 patients with acute rejection within the first week post-transplant, and 22 patients without rejection for at least 6 months post-transplant. Thirty-three transplant patients were clinically stable 3 months after kidney transplantation. A total of 183 probe sets representing 160 genes were found to be statistically significant and consistently differentially expressed between AR and NR subjects (Table 2). exist Figure 10 The sequences represented by the probe sets are shown in . Samples from subjects with acute rejection within the first week post-transplant were clustered together and separated from samples from patients without rejection.
[0289] Using the set of nucleic acid markers listed in Table 5, the experimental subjects were classified into rejecters (AR) or non-rejectors (NR) ( Figure 1A -C).
[0290] In contrast, an inde...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com