Method for calculating and simulating protein-protein docking
A protein and protein complex technology, applied in computing, special data processing applications, instruments, etc., can solve problems such as inability to distinguish between natural and wrong binding modes, and achieve the effect of improving screening efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0026] The method of the present invention is described in detail below with reference to the accompanying drawings.
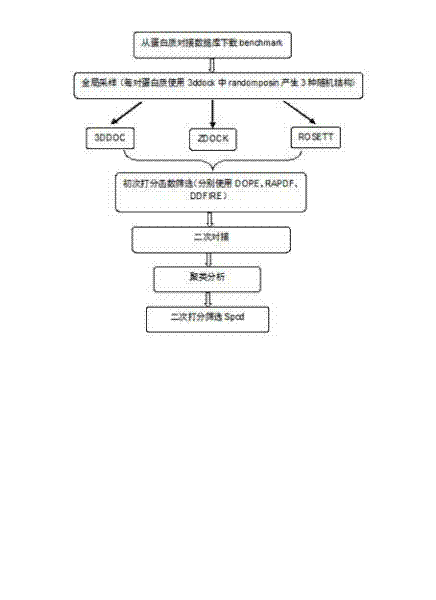
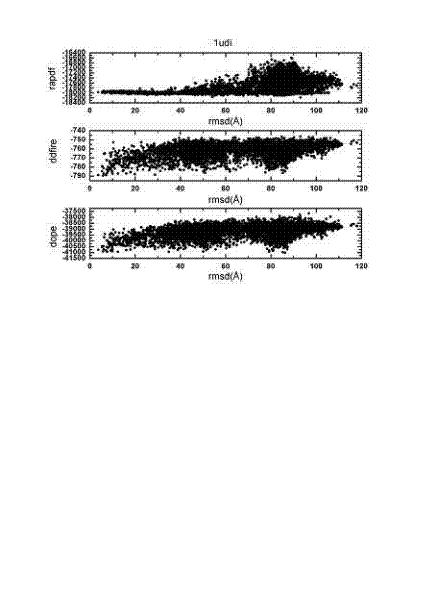
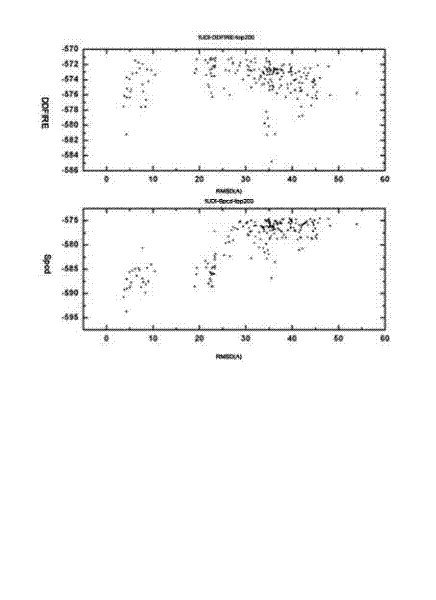
[0027] A computational simulation method for protein-protein docking, the idea of the present invention is as follows: We combine several different molecular docking methods, and after screening with different scoring functions, select a certain number of protein complexes, and then perform a new round of Molecular docking, the docked complexes were then subjected to cluster analysis based on structural similarity. Clustering analysis can group compounds with similar structures into one category, and we select the groups with the most conformations in the same group of complexes to use the scoring function we designed for screening. The scoring function combines the scoring function of ENDES and the scoring function of DDFIRE. The scoring function of ENDES takes into account protein structure propensity and protein evolution information. DDFIRE is a scoring...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com