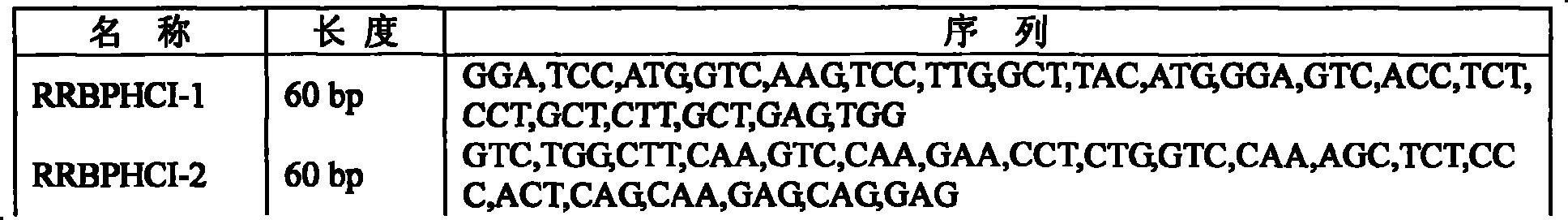Family shuffling technology system for modifying multiple genes of dihydroxyl dioxygenase
A technology of dihydroxydioxygenase and hydroxydioxygenase, which is applied in plant gene improvement, recombinant DNA technology, genetic engineering and other directions, can solve the problems of difficult target shape, low mutation rate and small mutation potential.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Embodiment 1: PTDS method synthesizes dihydroxydioxygenase gene PSBPHCI
[0026] (1) Test method:
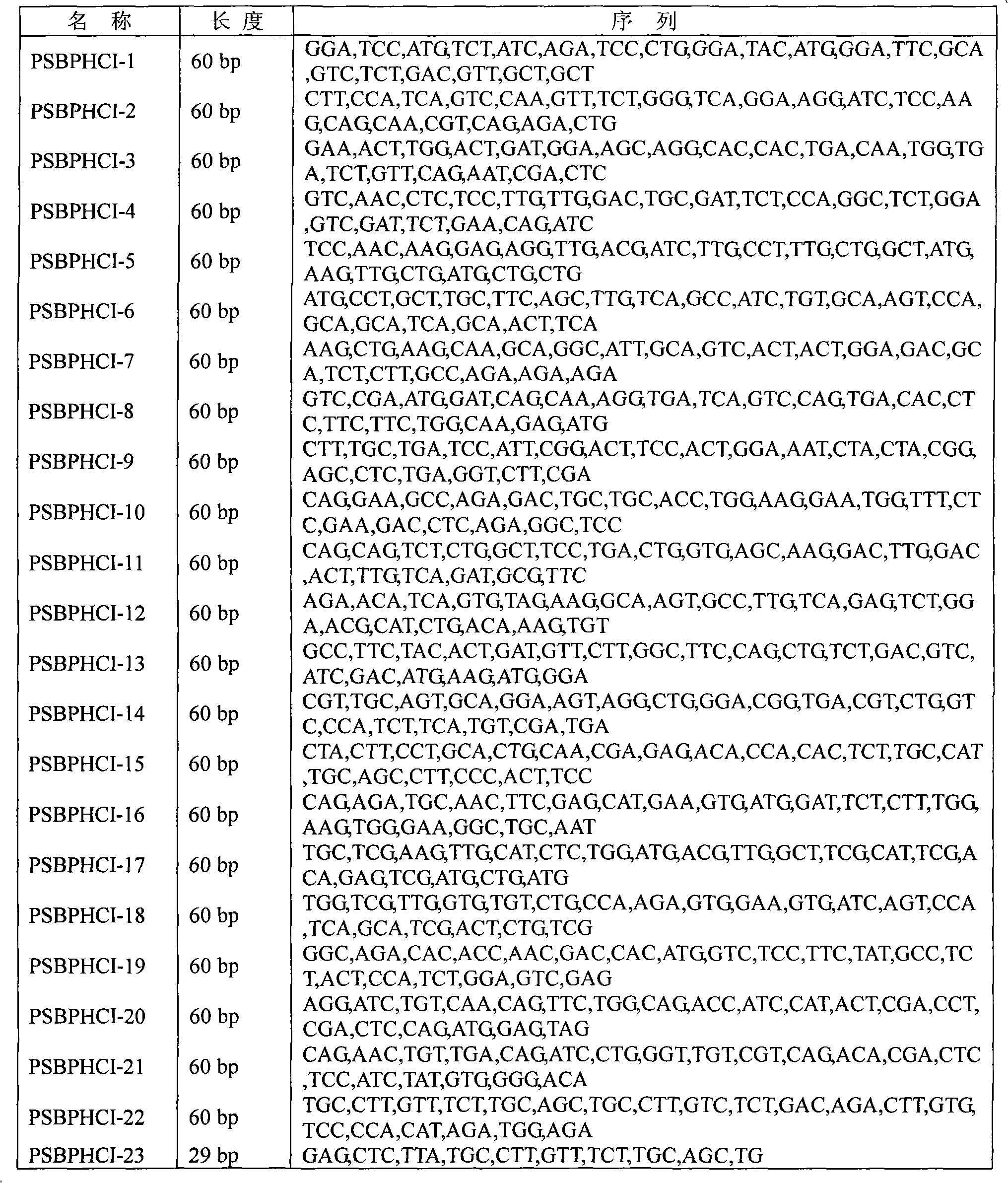
[0027] 1. The gene synthesis of the present invention adopts the PTDS method [Xiong'aisheng et al., 2004, Nucl Acids Res 32, e98], and a total of 23 primers are designed for gene synthesis, see Table 1.
[0028] Amplified Fragment 1: In a 50 μl reaction system, the amount of inner primers (PSBPHCI2-PSBPHCI11) was 1.5 pmol, and the amount of outer primers (PSBPHCI1, PSBPHCI12) was 30 pmol. The amplification conditions were: 94°C preheating for 10 minutes; 94°C , 30s; 52°C, 30s; 72°C, 30s; 30 cycles; finally 72°C extension for 10min. The Taq DNA polymerase used was pyrobest taq enzyme (Takara Company Dalian) to amplify the target gene.
[0029] Amplified Fragment 2: In a 50 μl reaction system, the amount of inner primers (PSBPHCI14-PSBPHCI22) added is 1.5 pmol, and the amount of outer primers (PSBPHCI13, PSBPHCI23) added is 30 pmol. The amplification conditions are: 94°C ...
Embodiment 2
[0036] Embodiment 2: PTDS method synthesizes dihydroxydioxygenase gene RRBPHCI
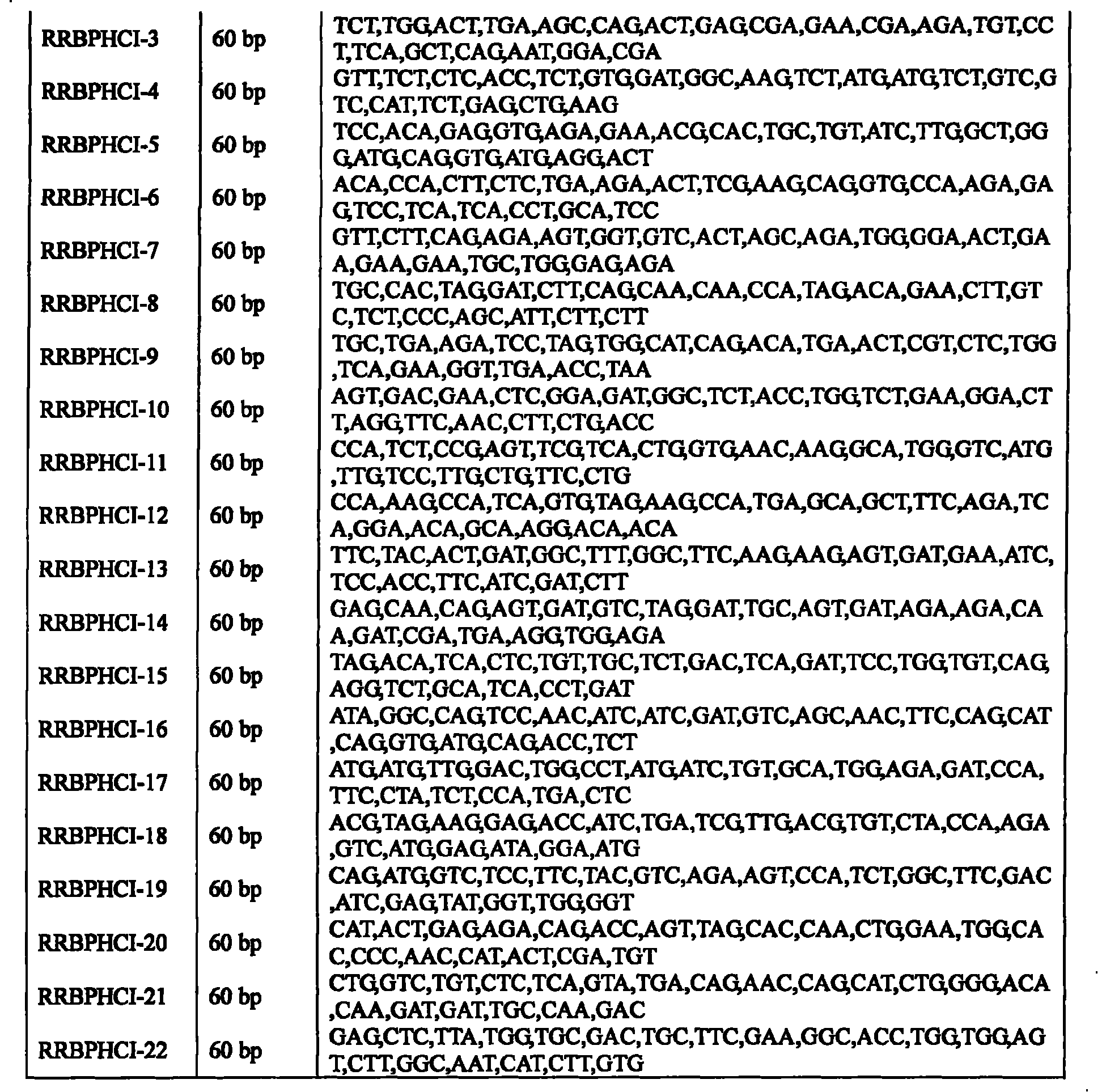
[0037] The gene synthesis of the present invention adopts the PTDS method [Xiong'aisheng et al., 2004, Nucl Acids Res32, e98], and a total of 22 primers are designed for gene synthesis, and the length of each primer is 60 bp. See Table 2. Both ends of the primers were respectively introduced with Bam H I and Sac I enzyme cutting sites.
[0038] Wherein the amplified fragment 1 (Fragment 1): in the 50 μ l reaction system, the addition amount of inner primers (RRBPHCI-2~RRBPHCI-9) is 1.5 pmol, and the addition amount of outer primers (RRBPHCI-1 and RRBPHCI-10) is 30 pmol, The amplification conditions were: preheating at 94°C for 10 min; 94°C for 30 s; 52°C for 30 s; 72°C for 30 s; 30 cycles; and finally extension at 72°C for 10 min. The TaqDNA polymerase used was pyrobest taq enzyme (Takara Company Dalian) to amplify the target gene.
[0039] Amplified Fragment 2 (Fragment 1): In a 50 μl reaction...
Embodiment 3
[0047] Embodiment 3: PTDS method synthesizes dihydroxydioxygenase gene MTBPHCI
[0048] The gene synthesis of the present invention adopts the PTDS method [Xiong et al., 2004, Nucl Acids Res 32, e98], and a total of 23 primers are designed for gene synthesis, wherein the length of the primers MTBPHCI-1 to MTBPHCI-22 is 60bp, and the primer MTBPHCI-23 The length is 35bp. See Table 3. Both ends of the primers were introduced with BamHI and SacI enzyme cutting sites.
[0049] Amplified Fragment 1 (Fragment 1): In a 50 μl reaction system, the amount of inner primers (MTBPHCI-2~MTBPHCI-9) was 1.5 pmol, and the amount of outer primers (MTBPHCI-1 and MTBPHCI-10) was 30 pmol. The amplification conditions were: preheating at 94°C for 10 min; 94°C for 30 s; 52°C for 30 s; 72°C for 30 s; 30 cycles; and finally extension at 72°C for 10 min. The TaqDNA polymerase used was pyrobesttaq enzyme (Takara Company Dalian) to amplify the target gene.
[0050] Amplified Fragment 2 (Fragment 1): ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com