Method and system for accelerating large-scale protein identification by using suffix array
A technology of protein identification and suffix array, applied in the field of suffix array to accelerate large-scale protein identification, method and system, can solve the problems of large inverted index time and space, inconvenient use, slow database search speed, etc., to improve identification Speed, less time and space, effects of reduced computation time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0058] The present invention will be described in detail below in conjunction with the accompanying drawings and specific embodiments, but not as a limitation of the present invention.
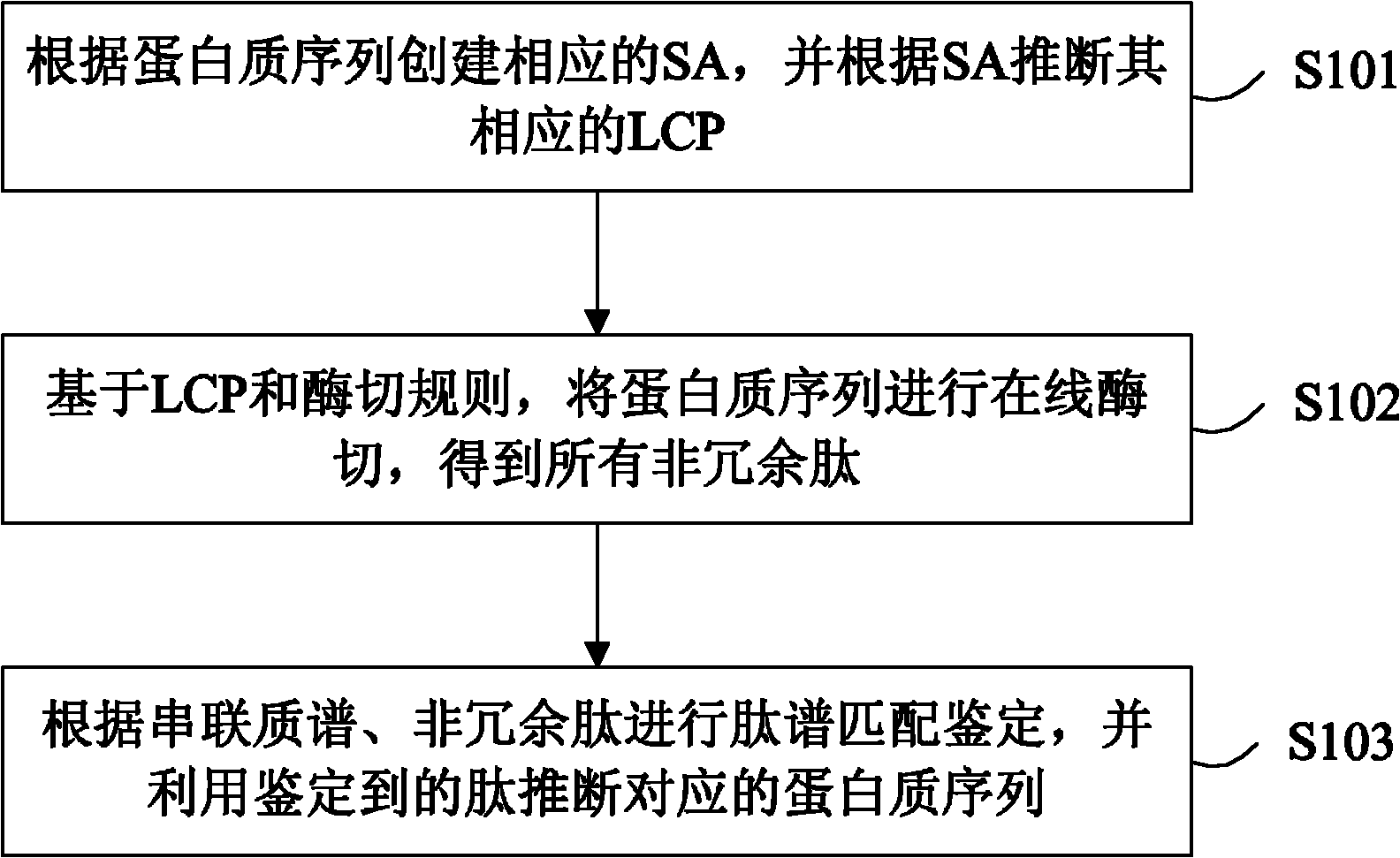
[0059] Such as figure 1 Shown is a flow chart of the method for accelerating large-scale protein identification using suffix arrays in the present invention, which is mainly used in database search methods based on tandem mass spectrometry in large-scale protein identification. The concrete steps of this method process include:
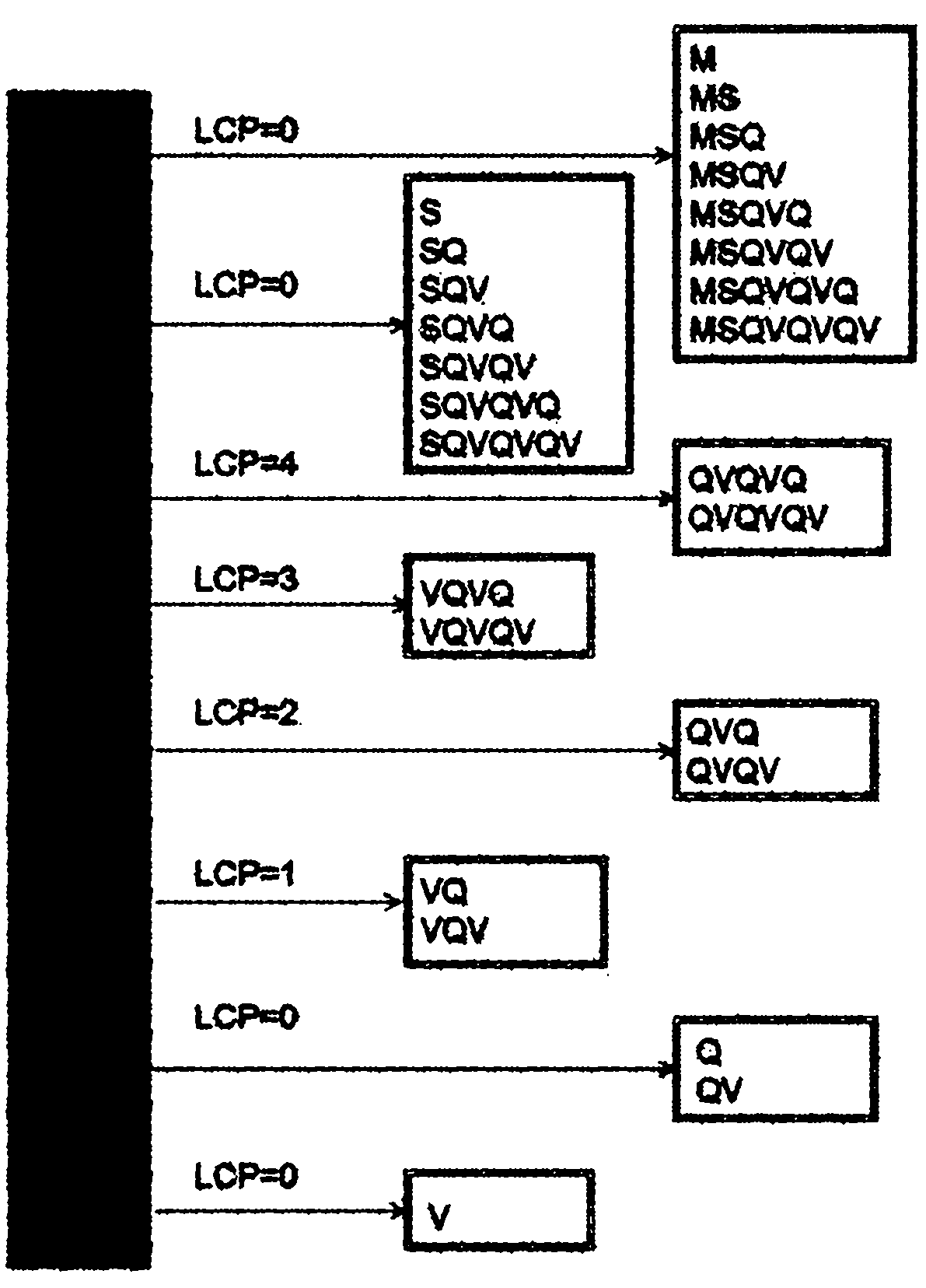
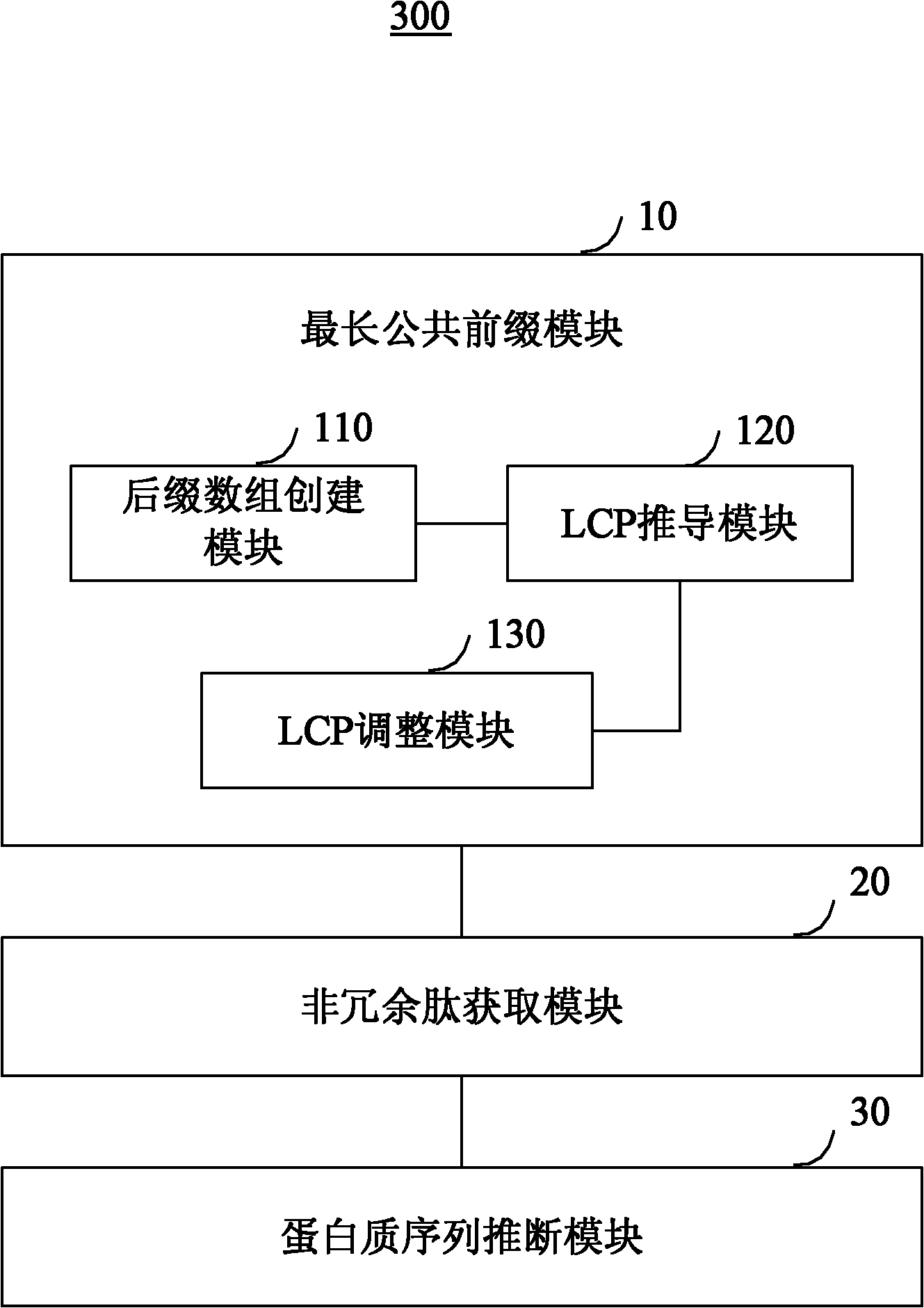
[0060] Step 101, create a corresponding suffix array SA according to the protein sequence in the database, and deduce the longest common prefix LCP corresponding to the protein sequence according to SA;
[0061] Step 102, based on LCP and digestion rules, the protein sequence is digested online to obtain all non-redundant peptides;
[0062] In step 103, the peptide spectrum matching identification is carried out according to the tandem mass spectrum and the non-redu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com