Method for detecting nucleotide sequence by using poly(methyl acrylic pyrene-poly (methyl) acrylic dimethylamine ethyl ester copolymer, and product thereof
A technology of dimethylamine ethyl acrylate and polypyrene methyl acrylate, which is applied in the field of functional polymers, can solve the problem of reducing the thermal stability of the hybrid double-chain of fluorescent probes and targeting molecules, complicated preparation of labeled fluorescent probes, Reduce the detection accuracy of fluorescent probes and other problems, and achieve the effect of simple and easy synthesis route, reduced fluorescence intensity, good selectivity and sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
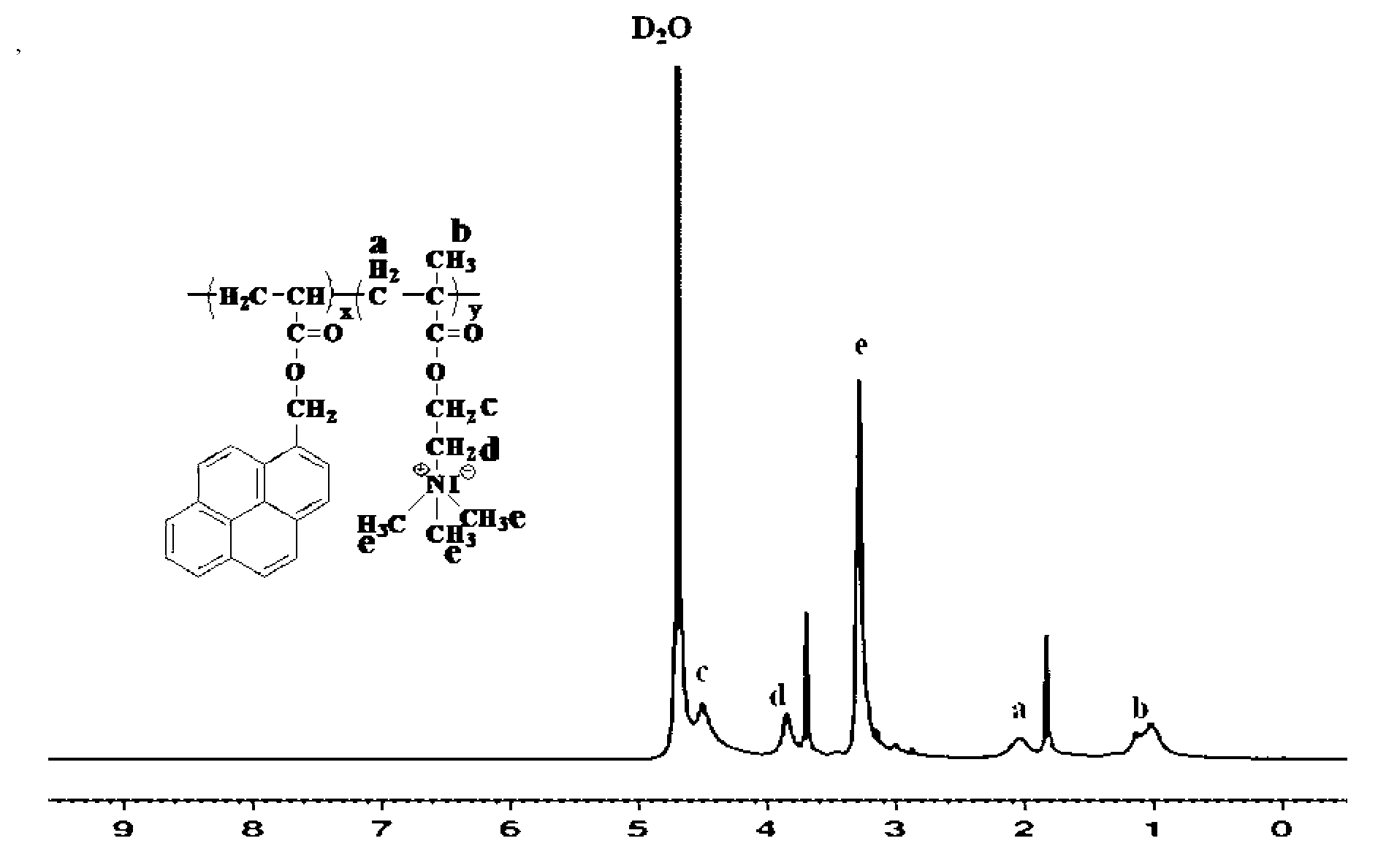
[0033] Add 2ml of dry THF solvent to a 25ml clean single-necked round bottom flask, then add 0.3mmol copolymer and 0.9mmol methyl iodide respectively, and react at room temperature for 24 hours. After the reaction, filter the solvent THF, and then Extract the product with a Soxhlet extractor, wherein the extractant is tetrahydrofuran, and the extraction time is 12 hours, and finally the final fluorescent polyelectrolyte (P(DMAEMA + -co-Py)) was dried in a vacuum oven at 70°C for 12 hours.
example 2
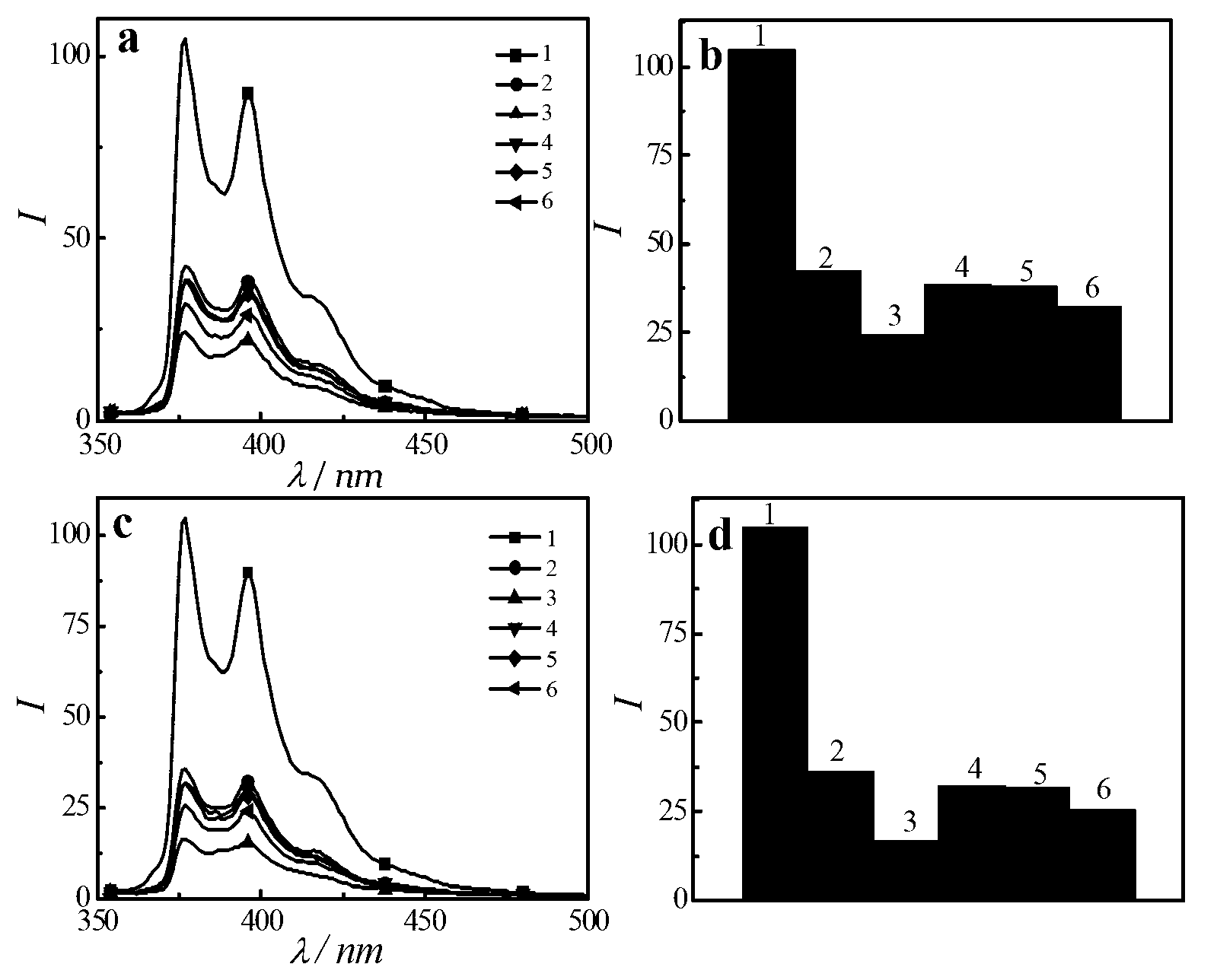
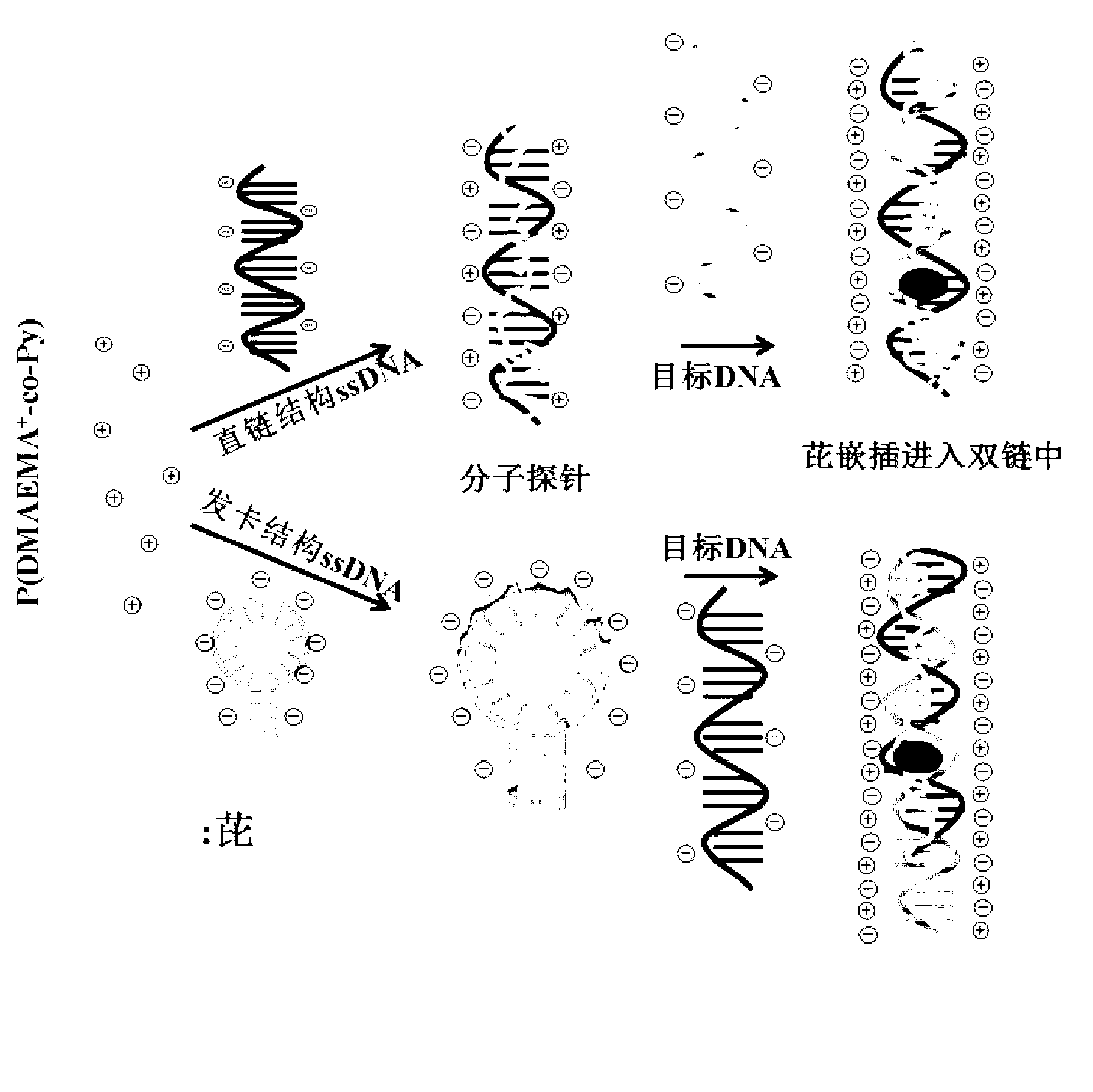
[0035] (1) Sample preparation: Weigh an appropriate amount of fluorescent polyelectrolyte P(DMAEMA + -co-Py) was dissolved in buffer PBS to make its original concentration 1×10 -5 M, use 1ml of buffer to dilute each tube containing 1od of DNA1, DNA2, DNA3, DNAa, DNAc, DNAt, wherein DNA2 is a hairpin structure, DNA1, DNA3, DNAa, DNAc and DNAt are linear structures, and DNA2 is partially complementary to DNA1, and completely non-complementary to DNAa, DNAc, and DNAt; DNA1 is completely complementary to DNA3, and completely non-complementary to DNAa, DNAc, and DNAt. Then take appropriate amount of fluorescent polyelectrolyte buffer and DNA buffer, and finally make P(DMAEMA + -co-Py), P(DMAEMA + -co-Py)+DNA1,P(DMAEMA + -co-Py)+DNA1+DNA3, P(DMAEMA + -co-Py)+DNA1+DNAa, P(DMAEMA + -co-Py)+DNA1+DNAc, P(DMAEMA + -co-Py)+DNA1+DNAt, P(DMAEMA + -co-Py)+DNA2,P(DMAEMA + -co-Py)+DNA2+DNA1, P(DMAEMA + -co-Py)+DNA2+DNAa, P(DMAEMA + -co-Py)+DNA2+DNAc, P(DMAEMA + -co-Py)+DNA2+DNAt11 s...
example 3
[0039] (1) Sample preparation: Weigh an appropriate amount of fluorescent polyelectrolyte P(DMAEMA + -co-Py) was dissolved in buffer PBS to make its original concentration 1×10 -5 M, use 1ml of buffer to dilute DNA1, DNA2, and DNA3 containing 1od in each tube, where DNA2 has a hairpin structure, DNA1, and DNA3 are linear structures, and DNA2 is partially complementary to DNA1, and DNA1 is completely complementary to DNA3. Then take appropriate amount of fluorescent polyelectrolyte buffer and DNA buffer, and finally make P(DMAEMA + -co-Py)+DNA1+DNA3, DNA1+DNA3, P(DMAEMA + -co-Py)+DNA2+DNA1, DNA2+DNA14 sample solutions, wherein in each sample solution, the concentration of fluorescent polyelectrolyte remains 4.0×10 -6 M, the concentration of DNA was kept at 1×10 -4 M.
[0040] (2) Sample heat treatment: Put the sample prepared in (1) into a 90°C oven for heat treatment for 5 minutes, and then quickly move to a 40°C oven for half an hour.
[0041] Finally, circular dichroism...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap