Typing method for multicolor fluorescence composite detection of 20 X-SNP sites
A technology of composite detection and typing method, applied in the direction of biochemical equipment and methods, microbial measurement/testing, etc., to achieve the effect of improving the success rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
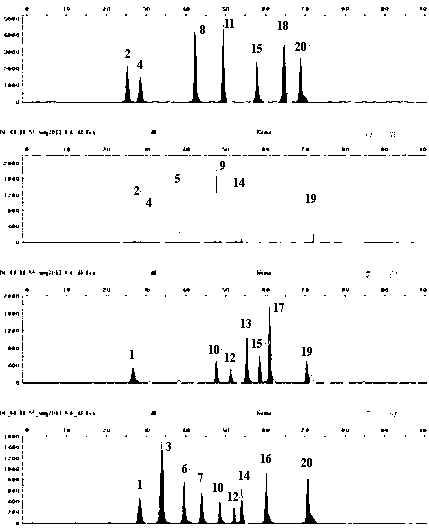
Image
Examples
Embodiment 1
[0028] (1) Extract the DNA of a woman to obtain a DNA solution;
[0029] (2) Amplification: Add the DNA solution obtained in step (1) to the amplification system, and then compound and amplify the DNA fragments of 20 X-SNP sites simultaneously in the amplification instrument.
[0030] The amplification system includes: PCR buffer solution, MgCl 2 , dNTPs, PCR primers, Taq DNA polymerase, template DNA; the concentrations or contents of the amplification system components are shown in Table 3:
[0031] Table 3 Concentration or content of amplification system components
[0032] Reagent volume 10×PCR buffer solution 2.5μL Mg 2+ (25mM) 5μL dNTPs (10mM) 1.5μL PCR primers (see Table 3 for concentration) 2.8μL Taq DNA polymerase (1U / μL) 1.5μL Template DNA (1-10ng / μL) 1μL Deionized water 10.7μL
[0033] The concentrations of PCR primers and extension primers are shown in Table 4.
[0034] Table 4 Concentrations of PC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com