Method for detecting differentially-expressed protein
A differential expression and protein technology, applied in the field of quantitative proteomics, can solve the problems of accuracy evaluation of quantitative results without protein, inaccurate qualitative and quantitative results, labor-consuming and material resources, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
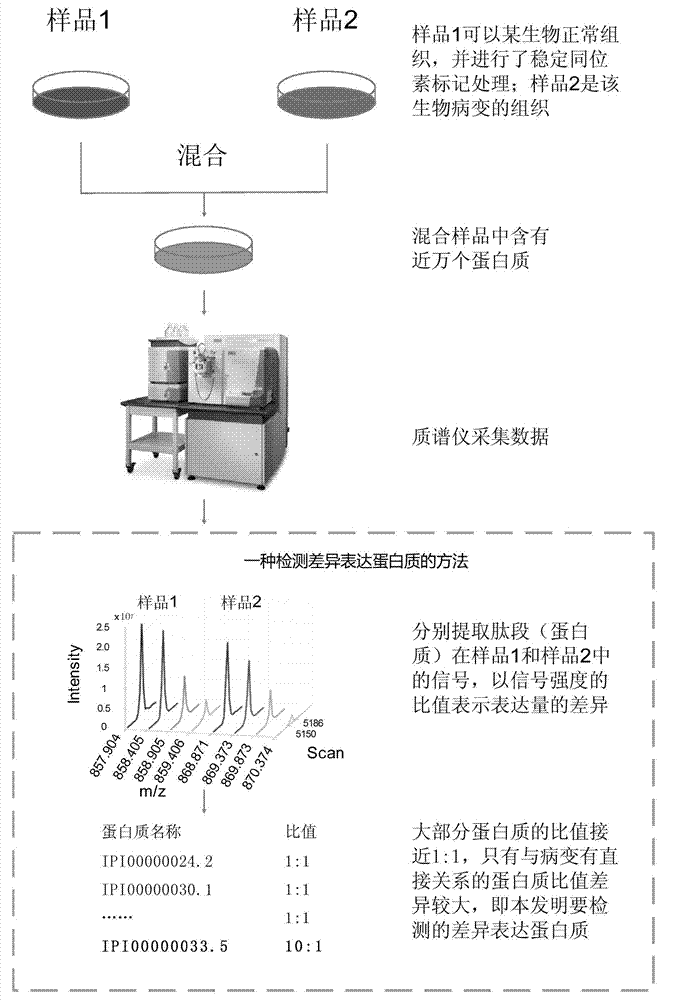
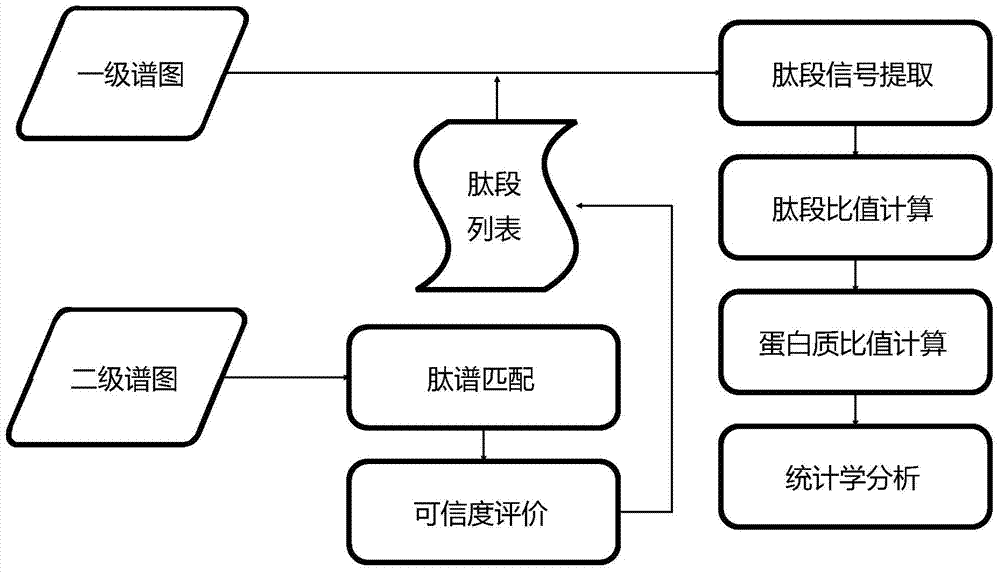
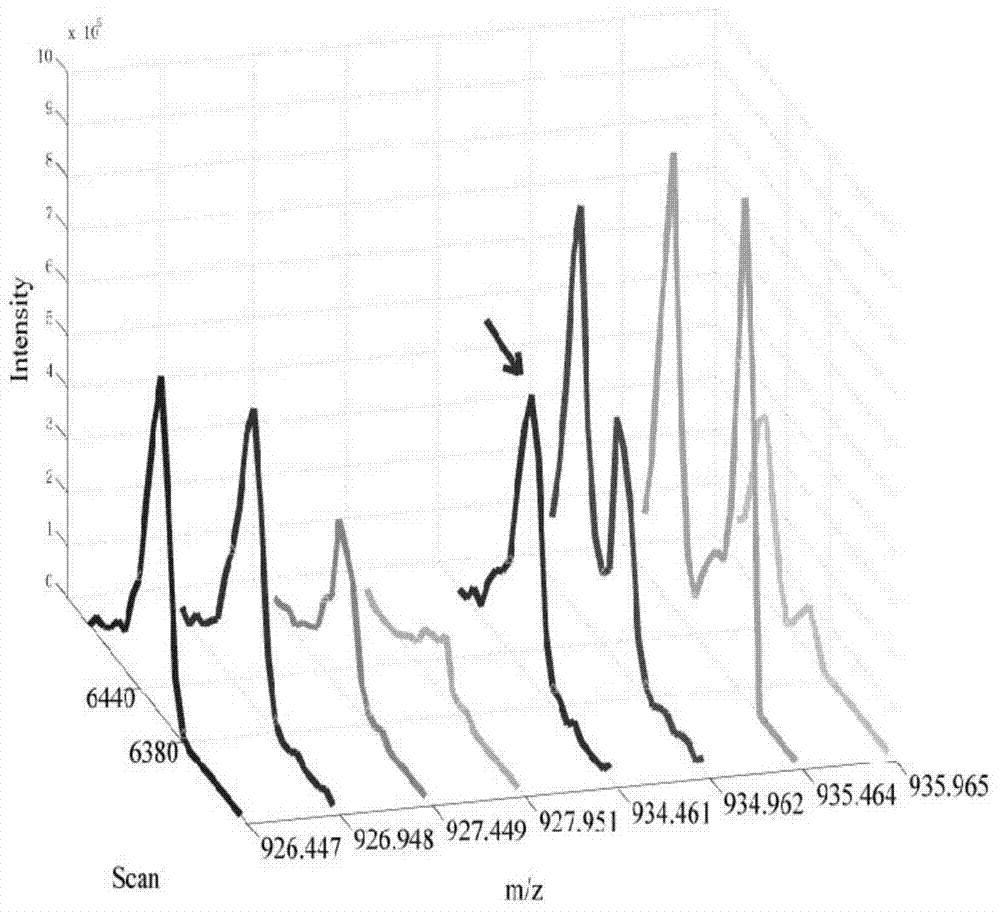
[0072] The present invention detects which are differentially expressed by analyzing the data of proteins collected by a mass spectrometer in different samples, specifically, the relative quantification of markers and non-markers based on primary spectrum information in quantitative proteomics Data analysis, according to the ratio of the mass spectrometry signal intensity corresponding to a protein in two or more biological samples to determine whether it is a differentially expressed protein.
[0073] Such as figure 1 Shown is the complete implementation process of the present invention, also is the experimental process of marking quantification, mainly comprises:
[0074] (1) Take two different biological samples for differential protein detection. Sample 1 is a normal tissue of a certain organism, which has been labeled with a stable isotope; sample 2 is a biological diseased tissue.
[0075] Use Stable Isotope Labeling by Amino acids in Cell culture (SILAC) or 15 N in vi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com