Indian truffle ssr primer and its cross-species screening method and application
An Indian truffle and product technology, applied in the field of molecular biology, can solve problems such as unseen population genetics research reports and difficulty in group sampling, and achieve the effects of shortening the analysis cycle, being convenient to use, and improving research efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Screening of SSR primers for Indian truffle (Tuber indicum):
[0060] 1. SSR primer design
[0061] ①Download the whole genome sequence of Tuber melanosporum from NCBI (http: / / www.ncbi.nlm.nih.gov) database.
[0062] ② Use SSRHunter1.3 to search for simple repeat sequence information sites in the genome sequence.
[0063] ③ Select the sequence with 2 to 6 nucleotide repeat motifs at the information site as the target sequence, use the software Primer Premier5.0 to design SSR primers, and use Oligo6.65 to verify the parameters of the primers. Each parameter meets the following conditions: GC The content is 45%-60%; the annealing temperature is 50-60°C; the expected fragment length is between 100-300bp; the primer length is between 18-25bp.
[0064] ④ Select primer sequences that meet certain conditions for synthesis.
[0065] 2. Primer amplifiability detection
[0066] By repeatedly exploring the PCR reaction system and amplification conditions, the following steps w...
Embodiment 2
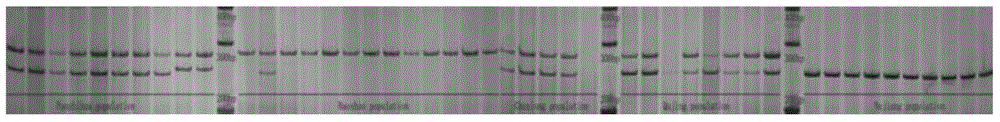
[0077] Genetic diversity analysis of Indian truffles based on SSR markers:
[0078] Using the SSR polymorphism primers screened in Example 1 to carry out genetic diversity analysis of Indian truffles, the steps are as follows:
[0079] The Indian truffle samples used in the examples were all collected from Yunnan and Sichuan, China.
[0080] 1. Analyze the source of the sample
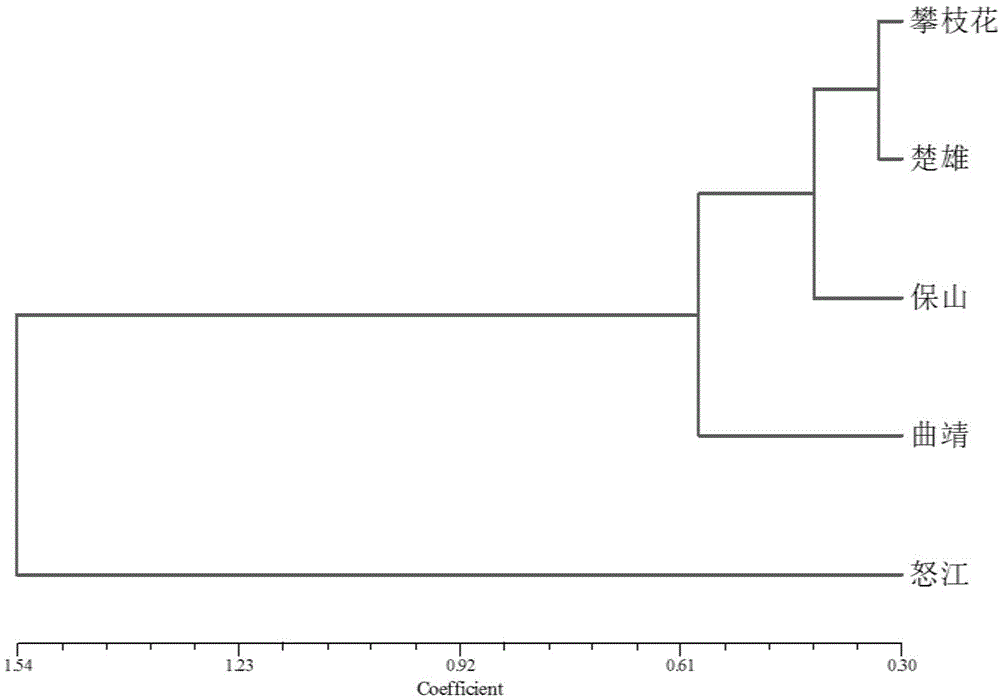
[0081] The samples used for the analysis were collected from wild Indian truffle populations in Yunnan and Sichuan. The specific geographic information is as follows: Qujing City, Yunnan (103°17′28.05″E, 26°24′46.84″N); Chuxiong Prefecture, Yunnan (101° 9′5.14″E, 25°35′54.49″N); Baoshan City, Yunnan Province (99°19′41.64″E, 25°12′18.34″N); Nujiang Prefecture, Yunnan Province (98°37′57.50″E, 28° 1′17.41″N); Panzhihua, Sichuan (101°37′58.18″E, 26°22′48.77″N).
[0082] 2. DNA extraction of Indian truffle samples
[0083] Use the improved 4×CTAB method to extract the total DNA of the sample, the specif...
Embodiment 3
[0108] Using SSR primers to construct the genetic map of Indian truffle, locate functional genes and establish germplasm resource bank.
[0109] Individuals of the same species of truffles with different geographical distributions usually show certain differences in phenotypic characteristics and taste attributes. Whether these differences are affected by environmental factors (symbiotic tree species, soil, etc.) or determined by genes is still controversial. SSR markers can analyze a large number of individuals with different geographical origins, thereby constructing a genetic map of Indian truffles, and locating genes that affect truffle ascocarp fruit shape and aroma, while certain genotypes are only in populations in specific regions If it exists, germplasm resource identification and preservation can be carried out for materials from different sources, and a corresponding germplasm resource bank can be established. For example, the 170bp gene in SSR24 only exists in the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com