Specific sequence and application of citrus sour rot germs
A technology specific to citrus sour rot, applied in the direction of microorganism-based methods, microbial measurement/inspection, biochemical equipment and methods, etc. Timely prevention of problems such as the urgent need and the rise of acid rot
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Obtaining the specific sequence of Solanum citri:
[0026] Our laboratory screened a citrus acid rot fungus, sequenced its whole genome, compared it with Geotrichum candidum genome and Geotrichum fermentans genome, and finally obtained the A specific sequence of Sour rot citri (shown in SEQ ID NO.1), the sequence exists specifically in Sour rot citri, so the sequence can be detected to determine the existence of Sour rot citri.
[0027] In the embodiment of the present invention, PCR primers are designed for this sequence for detection, and the primers are as follows: GTCGTCGTGTCCCCTGTTATGG and AAATGCTTGTGGGTTGCTCTT.
[0028]利用该引物可特异性的在柑橘酸腐病菌中扩增得到609bp的核苷酸序列,具体如下:GTCGTCGTGTCCCTGTTATGGTTTCTGGTAAAACTCTACCTTCGTTCAAGCCTTTCGATACTCATGCTCGTGCTGGTGGTTACATTTCTGGCCGTTTCTTTTCTGGTATCAAGCCTCAGGAATACTACTTCCACTGCATGGCTGGTCGTGAGGGTCTGATTGATACTGCCGTCAAGACTTCTAGATCCGGTTATCTGCAGAGATGTTTGATTAAGCAGCTCGAAGGTGTTCAGGTTAACTACGACAACTCCGTTCGTGACTCTGATGGTACCCTGGTTCAGTTTATGTACGGTGGTGATGCTGTCGATGTCA...
Embodiment 2
[0030] The application of the specific sequence of citrus sour rot bacteria comprises the following steps:
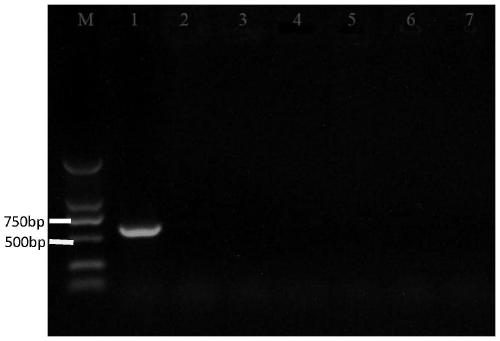
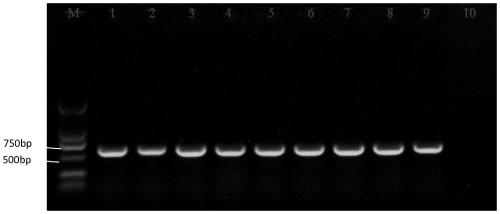
[0031] This embodiment utilizes the primers designed in Example 1 to amplify different bacteria belonging to the genus Geotrichum to investigate the specificity of the specific sequence provided by the present invention, as follows:
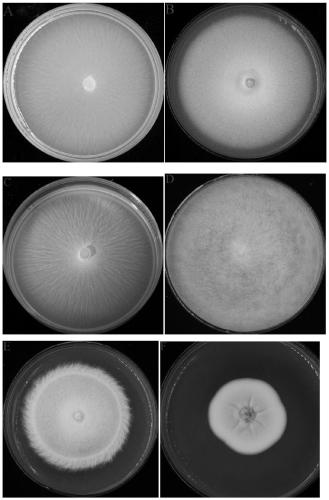
[0032] 1. Activation of different fungi: Take out Geotrichum citri-aurantii, Geotrichum sp., Geotrichum ludwigii, Geotrichum suaveolens from the -80°C ultra-low temperature refrigerator ), Geotrichum candidum and Geotrichum candidum 6 strains of Geotrichum fungi ( figure 1 ), and randomly selected 8 mutants from the citrus acid rot mutant library (Zheng Feng, 2017) and put them in ice to melt, and took 2ul of the bacteria liquid and put it in a constant temperature incubator at 28°C for 3-4 sky.
[0033] 2. Extract DNA from different fungi: pick activated hyphae or spores in PDB medium, place them at 28°C, 180r / min, and culture in shake ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com