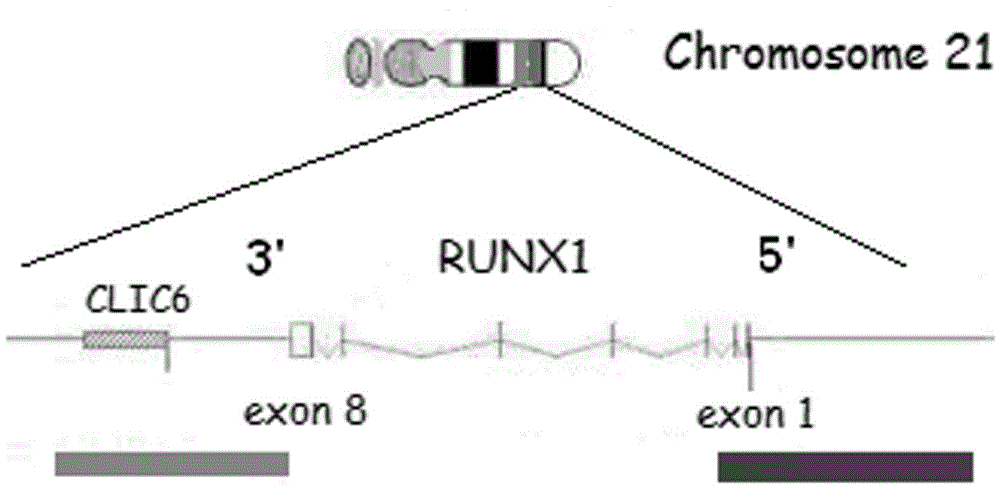
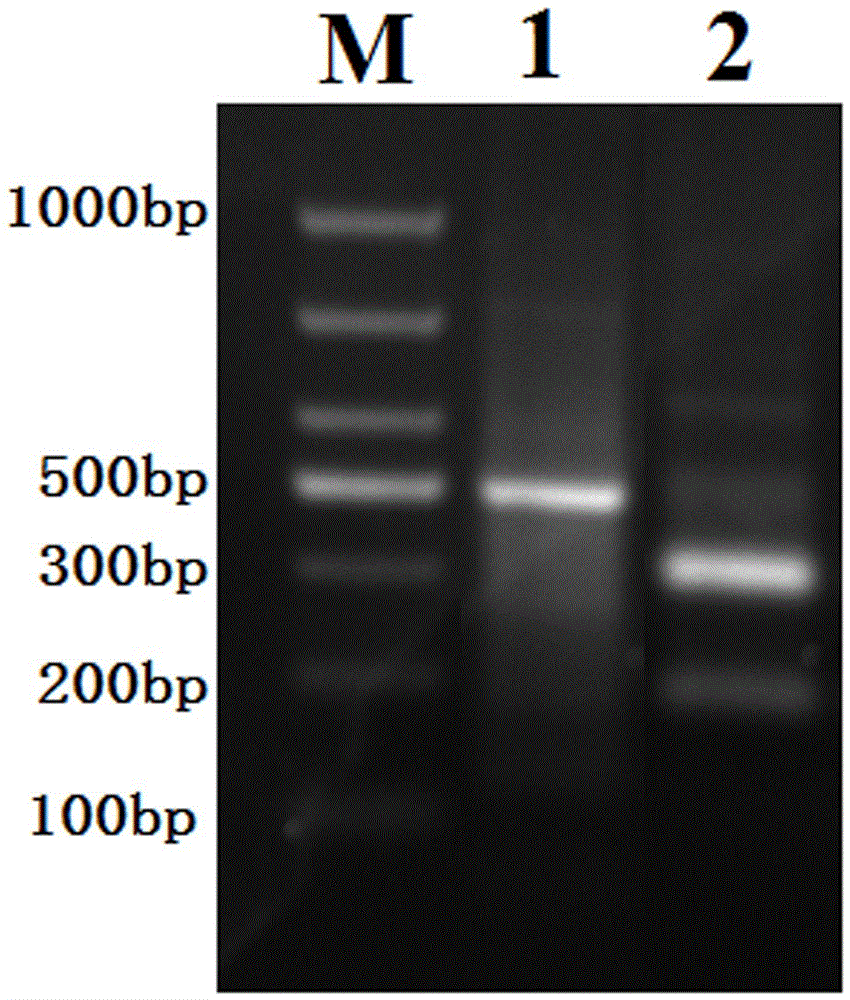
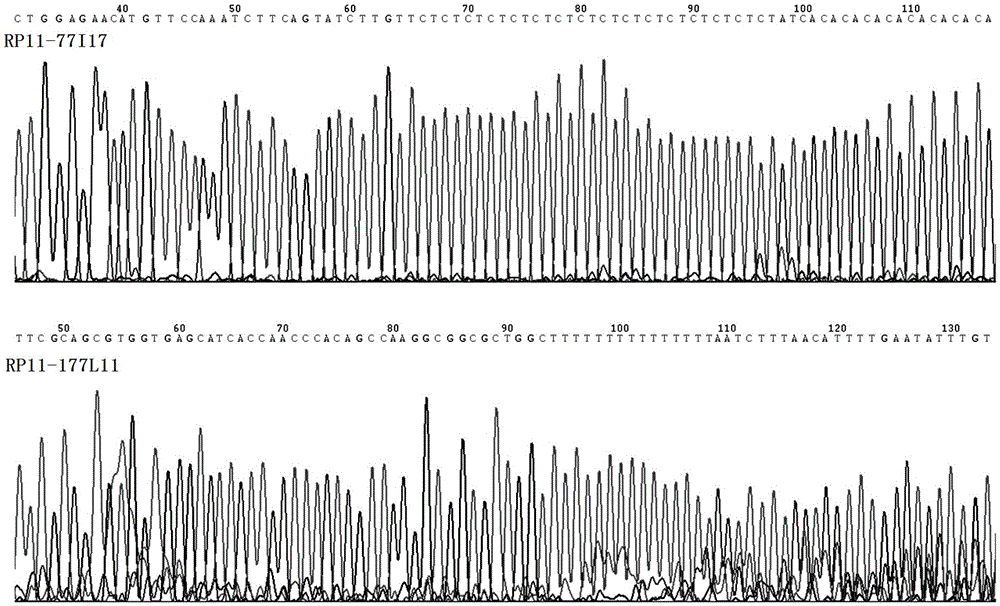
Runx1 gene fragmentation and copy number increase detection kit and preparation method thereof
A copy number increase and detection kit technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problem that it is not suitable for the screening of RUNX1 gene breaks, can not detect small chromosomal variation gene duplication, can not detect unknown Problems such as gene fracture and fusion, to achieve the effect of accurate identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0035] The present invention is further described below in conjunction with embodiment, but the present invention is not limited to the content of embodiment.
[0036] A RUNX1 gene fragmentation and copy number increase detection kit, which consists of the following components: Reagent I, Purification Reagent II, Precipitation Reagent III, Hybridization Buffer Reagent IV, the amount of each reagent in the kit is shown in Table 1, the amount of the listed reagents Amount for 10 probes:
[0037] Table 1
[0038]
[0039] Among them: the components of reagent I and the concentration and dosage of each component are shown in Table 2,
[0040] Table 2
[0041]
[0042] The components of reagent II and the concentration and dosage of each component are shown in Table 3.
[0043] table 3
[0044]
[0045] The components of reagent III and the concentration and dosage of each component are shown in Table 4.
[0046] Table 4
[0047]
[0048] The components of reagent I...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com