High throughput single nucleotide polymorphism assay
A technology of oligonucleotides and fluorescent dyes, applied in the field of high-throughput single nucleotide polymorphism assays, can solve the problems of expensive, difficult and time-consuming high-throughput detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Example 1: DNA extraction and quantification
[0069]A method for detecting the HaAHASL1-A122(At)T mutant allele was developed using plants from a sunflower line previously characterized as homozygous for the HaAHASL1-A122(At)T mutant allele. In addition, a method to detect the HaAHASL1-A122(At)T wild-type allele was developed using another sunflower line previously characterized as homozygous for the HaAHASL1-A122(At)T wild-type allele. As a result, a HaAHASL1 method consisting of a homogeneous assay detection system for a PCR process utilizing FRET was developed. After the development of the HaAHASL1 method consisting of a homogeneous assay detection system for the PCR process using FRET, it was validated using previously genotyped sunflower lines.
[0070] Genomic DNA was extracted from leaf tissues of sunflower lines homozygous for the HaAHASL1-A122(At)T mutant allele and a sunflower line homozygous for the HaAHASL1 wild-type allele using the QiaGene DNeasy96 Plant...
Embodiment 2
[0071] Example 2: Primer Design
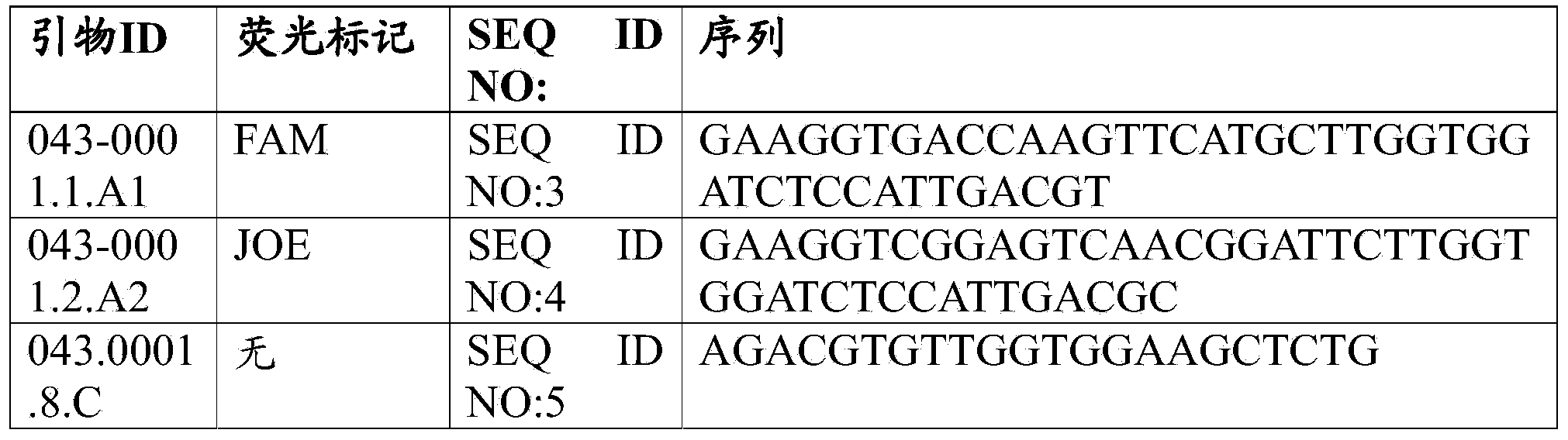
[0072] Three primers were manually designed based on the nucleotide sequence information of HaAHASL1 (SEQ ID NO:1) and HaAHASL1-A122(At)T mutant allele (SEQ ID NO:2) (Table 1).
[0073] A mutant allele detection common primer 043-0001.1.A1 (SEQ ID NO: 3) containing a tail at the 5' end was synthesized, and the sequence of the tail was the same as the fluorescently labeled primer of the KBiosciences reaction kit. KBiosciences Reaction Kit Fluorescently Labeled Primers are labeled with the fluorescent dye 5-carboxyfluorescein (FAM). This primer is specifically designed to amplify the HaAHASL1-A122(At)T mutant allele.
[0074] A wild-type allele detection common primer 043-0001.2.A2 (SEQ ID NO: 4) containing a tail at the 5' end that is the same as the fluorescently labeled primer of the KBiosciences reaction kit was synthesized. KBiosciences Reaction Kit Fluorescently Labeled Primers are labeled with the fluorescent dye 2’,7’-dimethoxy-4’,5’-d...
Embodiment 3
[0077] Example 3: HaAHASL1 single nucleotide polymorphism assay
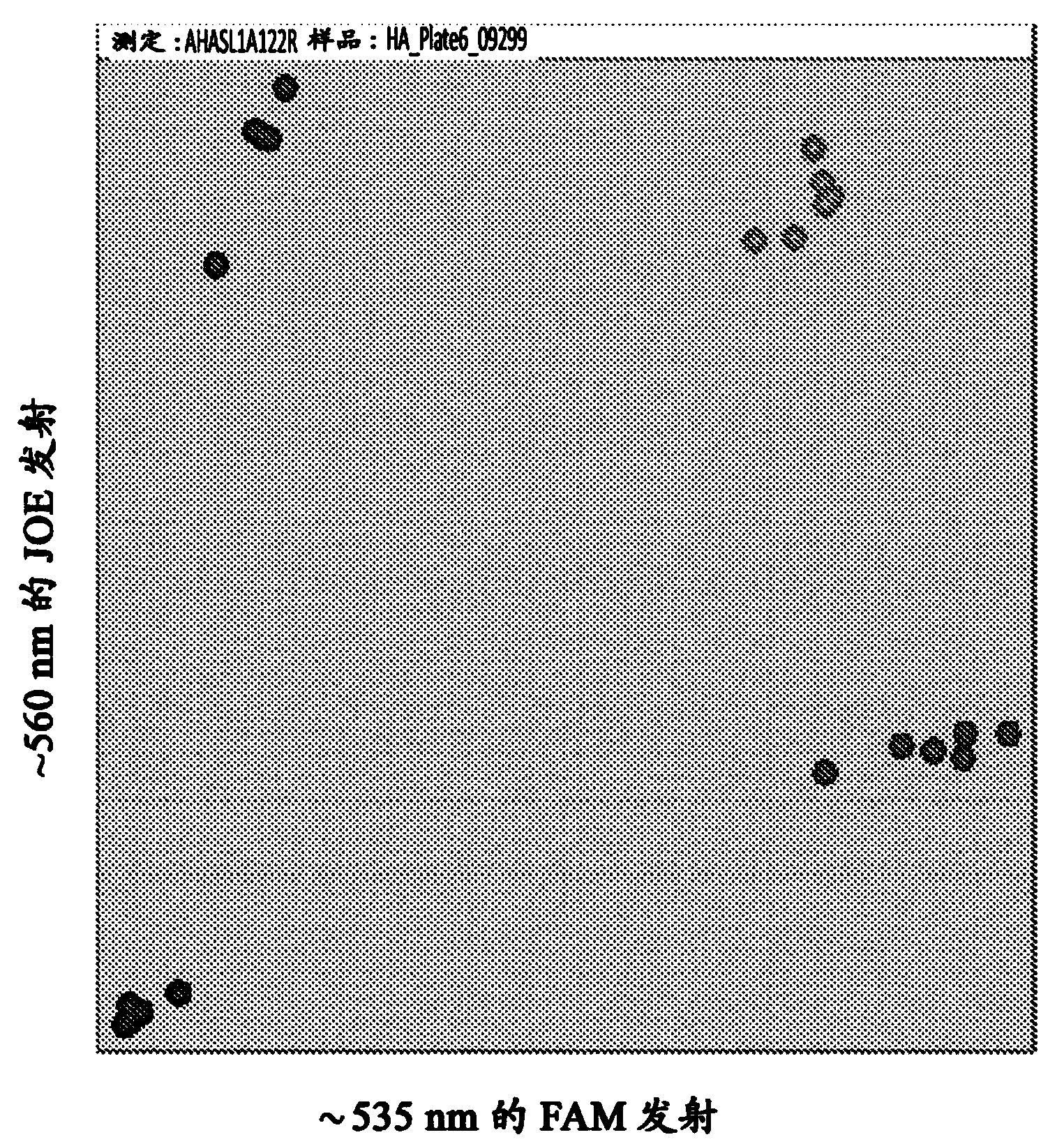
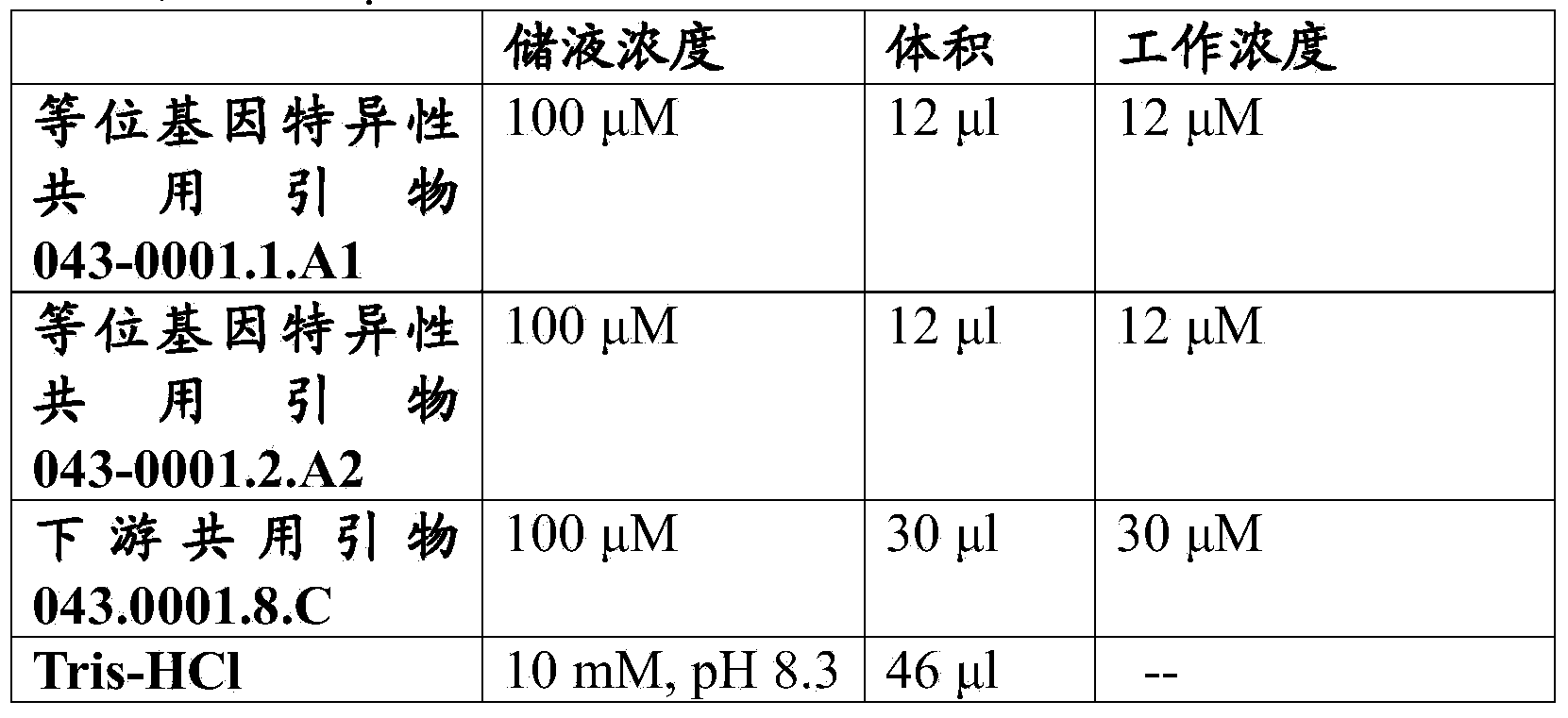
[0078] The three HaAHASL1 common primers were mixed to achieve the concentrations described in Table 2. The HaAHASL1 reaction cocktail and thermal cycling program are described in Tables 3 and 4, respectively. exist HaAHASL1 PCR reactions were performed in a 96-well plate format on a PCR System 9700 (Applied Biosystems, Carlsbad, CA). HaAHASL1 PCR reaction products were read on a fluorescent microplate reader using the following parameters: (1) FAM: excitation at about 485 nm; emission at about 535 nm; and (2) JOE: excitation at about 525 nm, emission at about 560 nm. Fluorescence readout data were saved in Microsoft Excel format and the data were plotted on the x-y axis. The FAM values are plotted on the X-axis and the JOE values are plotted on the Y-axis.
[0079] figure 1 The results of the assay are presented in . Samples known to be homozygous for the HaAHASL1 wild-type allele are clustered in th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com