Analysis method for single nucleotide polymorphism
A detection method and specific technology, applied in the field of in vitro diagnostic nucleic acid analysis and detection, can solve problems such as unknown mutations, affecting analysis results, differences between melting curve peaks and actual sample amplification results, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
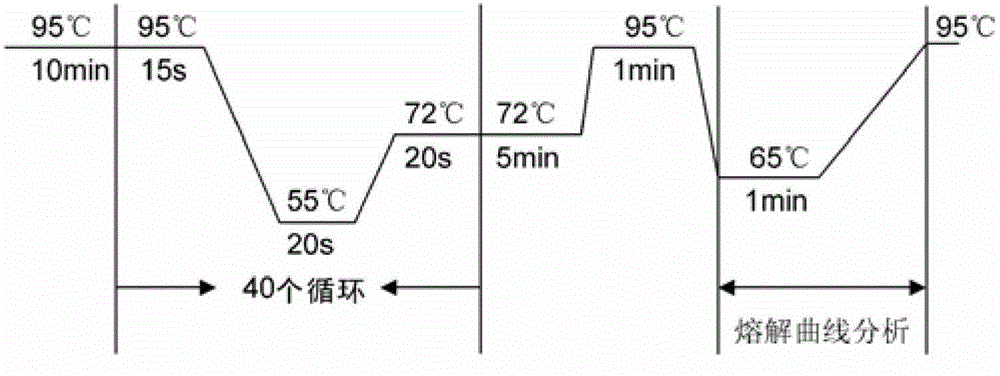
[0102] Embodiment 1, specific primer screening experiment
[0103] The purpose of the experiment: to introduce mutations, adjust the positions and bases of SNP sites and introduced mutation sites, and screen specific primers.
[0104] 1. Experimental materials
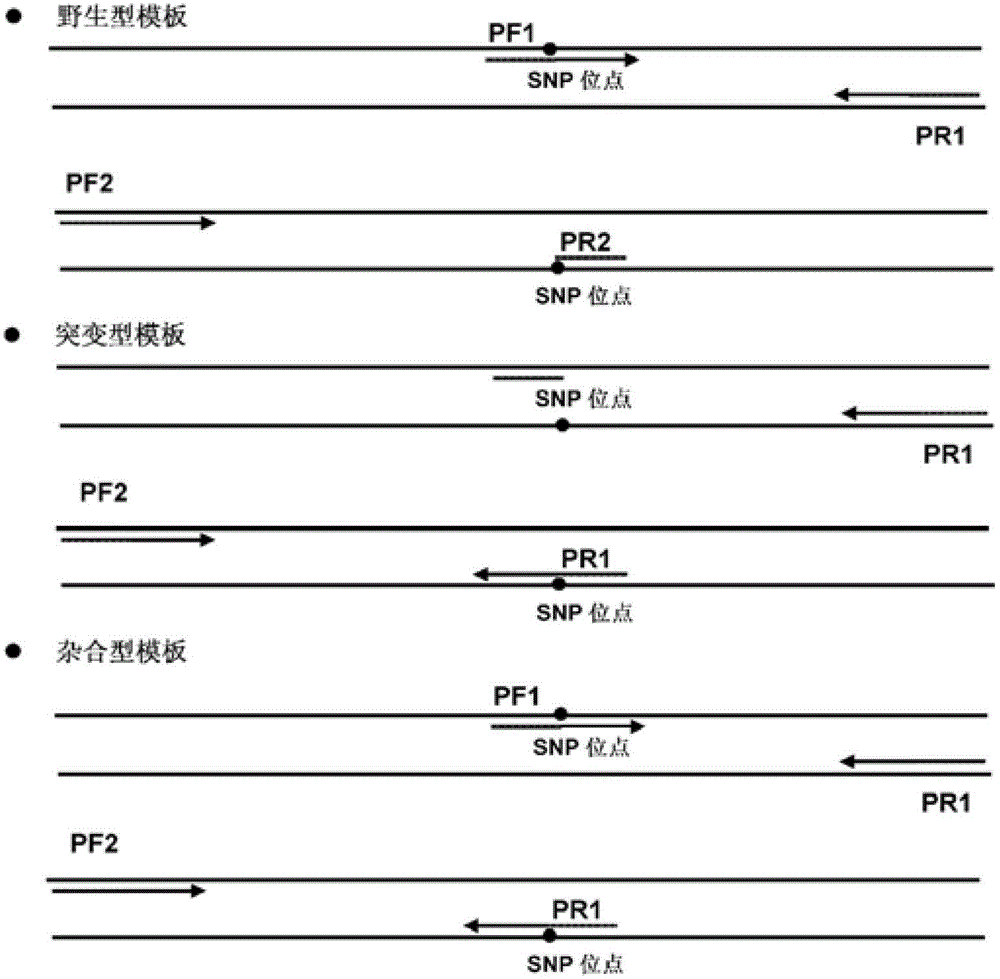
[0105] Primers were screened using wild-type, mutant and heterozygous samples to analyze primer sensitivity and specificity.
[0106] 2. Experimental grouping
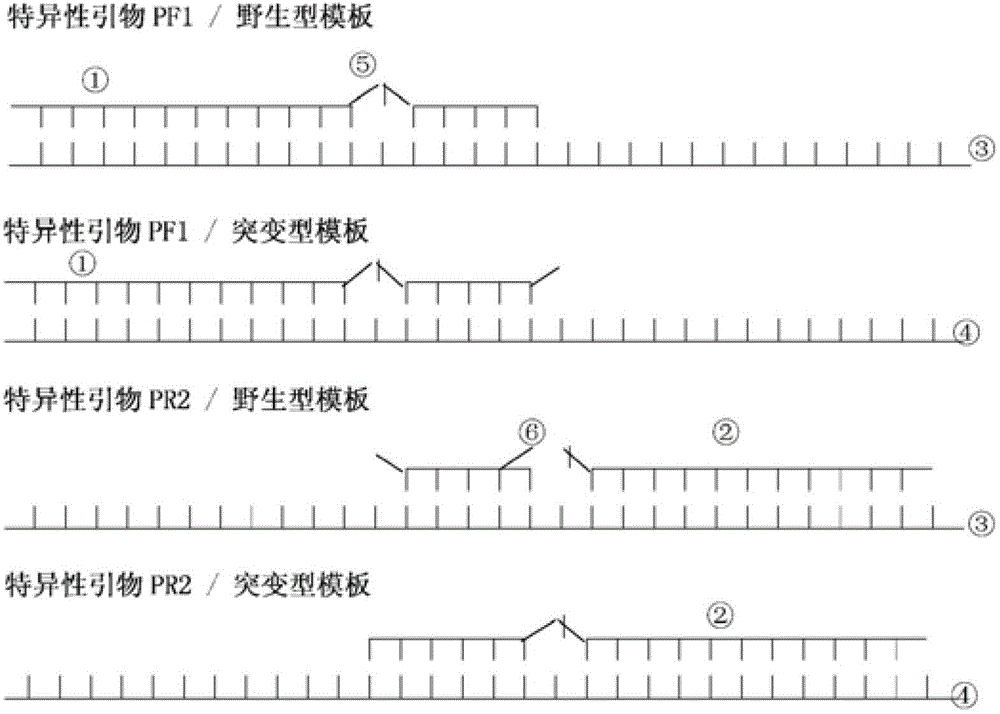
[0107] Group 1: primer set TPPF_WT_1 (specific primer, referred to as "special") and TP-2-R2 (common primer, referred to as "general");
[0108] Group 2: primer set TPPF_WT_2 (special) and TP-2-R2 (general);
[0109] Group three: primer set TP-2_W_F3 (special) and TP-2-R2 (general);
[0110] Group 4: primer set TP-2_W_F4 (special) and TP-2-R2 (general).
[0111] 3. Primer sequences and related information of PCR products
[0112] TPPF_WT_1: 5'-TGTATGATTTTATGCAGG TT C-3' (SEQ ID NO: 3);
[0113] TPPF_WT_2: 5'-TGTATGATTTTATGCAGG TT C-3' (SEQ ID NO:...
Embodiment 2
[0129] Example 2, common primers and multiple allele-specific primers PCR amplification melting curve analysis
[0130] Experimental purpose: To investigate the results of melting curve analysis of common primers and multiple allele-specific primers in PCR amplification.
[0131] 1. Experimental materials
[0132] The wild-type, mutant and heterozygous samples were used to investigate the melting curve analysis results of different primer sets.
[0133] 2. Experimental grouping
[0134] Common primer set: group 1: TPMT2-PF01 (common) and TPMT2-PR01 (common), group 2: TPMT2-PF02 (common) and TPMT2-PR02 (common);
[0135] Multiple allele-specific primer set: Group 3: TP-2_W_F3 (special) and TP-2-R2 (general), TP-2-F (general) and Set2-M R2 (special) (the ratio of primers has been adjusted) .
[0136] 3. Primer sequences and related information of PCR products
[0137] TPMT2-PF01: 5'-AAATGTATGATTTTATGCAGGTTT-3' (Tm=55.6°C) (SEQ ID NO:8);
[0138] TPMT2-PR01: 5'-CACACCAACTAC...
Embodiment 3
[0163] Example 3, Peripheral Primer Amplification, Effect on Multiple Allele Specific Primer PCR Amplification Melting Curve Analysis
[0164] The purpose of the experiment: To investigate the effect of peripheral primer amplification on the melting curve analysis of PCR amplification with multiple allele-specific primers.
[0165] 1. Experimental materials
[0166] Use wild-type, mutant and heterozygous samples to investigate the melting curve analysis results of different primer sets;
[0167] 2. Experimental grouping
[0168] Group 1: TP-2_W_F3 (special) (1ul), Set2-M R2 (special) (1ul) and TP-2-F (common) (0.1ul), TP-2-R2 (0.1ul) (common) ( Peripheral primers cannot effectively amplify); the concentration of common primers is 10 times lower than that of specific primers;
[0169] Group 2: TP-2_W_F3 (0.5ul) (special), Set2-M R2 (0.5ul) (special) and TP-2-F (common) (0.5ul), TP-2-R2 (0.5ul) (common ) (peripheral primers can effectively amplify).
[0170] 3. Information ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com