PKU (Phenylketnuria) gene detection kit
A gene detection and kit technology, applied in the field of kits for determining genotypes, can solve the problems of inability to determine mutation sites and mutation content, and achieve the effects of high accuracy, simplified operation process, and reduced operation steps.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] One, the information of 7 polymorphic sites on the PAH gene targeted by the present invention is as follows:
[0046]The PAH gene polymorphism sites selected by the present invention include: R111X(C→T), Y204C(A→G), R243Q(G→A), R252Q(G→A), R261Q(G→A), Y356X (C→A), R413P (G→C). The arrangement of each point is as follows figure 1 shown.
[0047] 2. The main components of the detection kit:
[0048] 1. Amplification primers for 7 polymorphic sites (R111X, Y204C, R243Q, R252Q, R261Q, Y356X, R413P):
[0049]
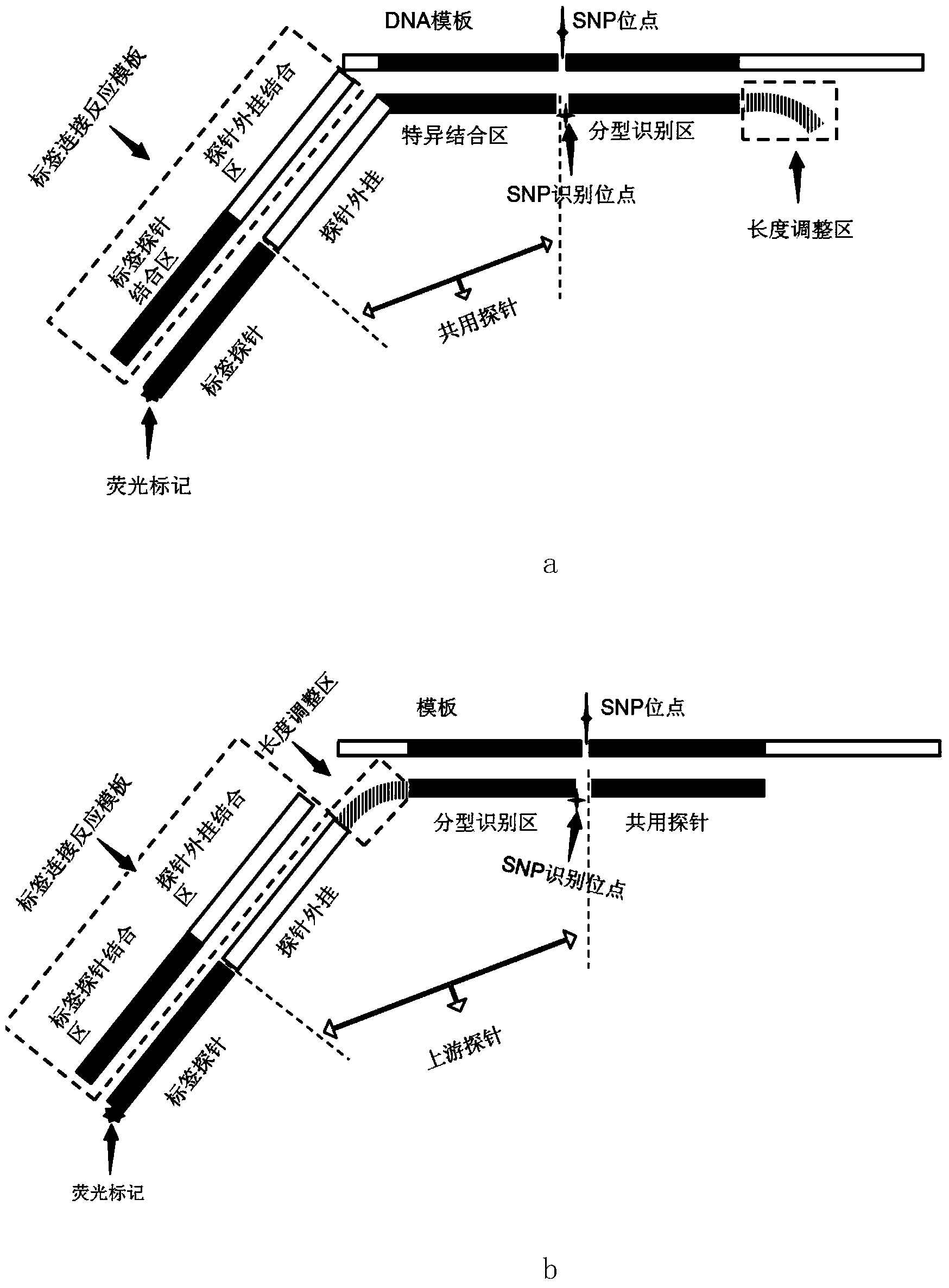
[0050] Among the above primers, F is an upstream primer, and R is a downstream primer. R243Q, R252Q, and R261Q are adjacent polymorphic sites, and a pair of amplification primers can amplify a DNA fragment containing these three polymorphic sites.
[0051] 2. Probes targeting polymorphic sites:
[0052]
[0053]
[0054] Among the above probes, F represents the common upstream common probe, W represents the wild-type downstream probe, and M represents t...
Embodiment 2
[0082] 1. Establishment of standard atlas
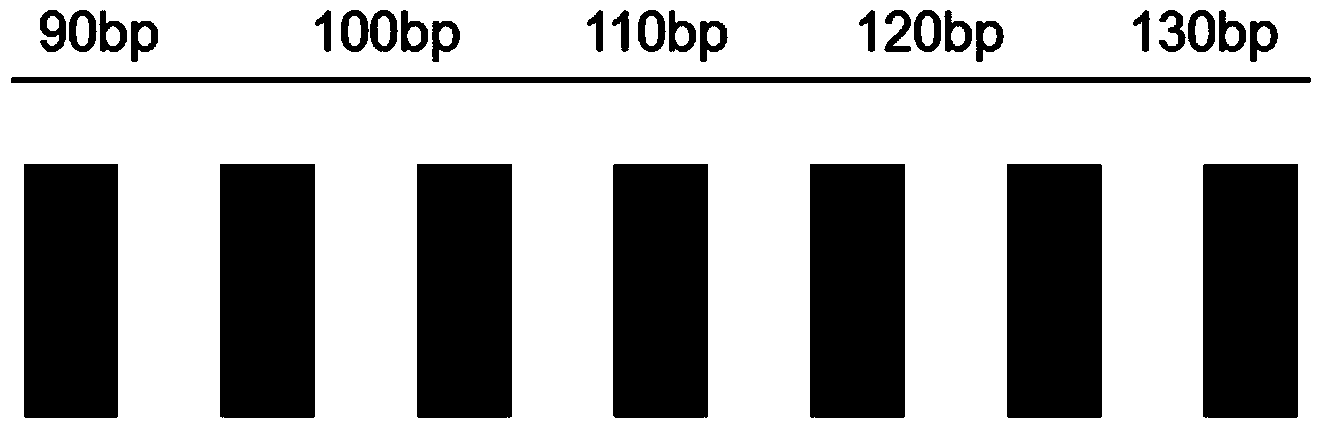
[0083] Use the kit and method of implementation 1 to detect 7 sites in the wild-type samples of the known PKU gene, and the detection map is shown in Figure 5 . Use the method of implementation 3 to detect 7 sites of known PKU gene mutation samples, and the detection map is shown in Figure 6 .
[0084] 2. Practical application examples
Embodiment 3
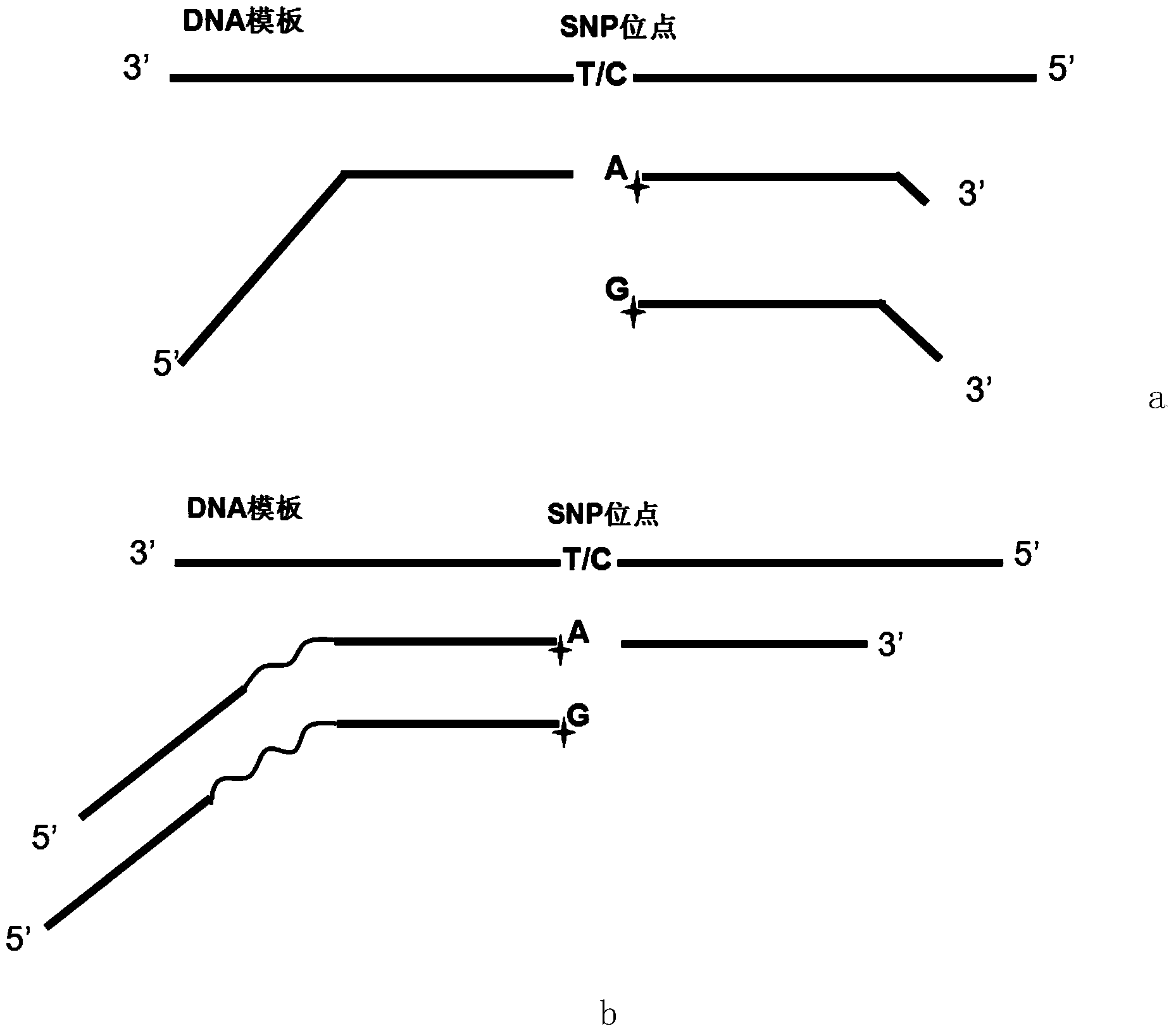
[0087] In order to improve the specificity of the ligase connection, the present invention artificially introduces a mismatch at the third base at the 5' end of the probe to increase the specificity of the probe connection. The principle of introducing mismatches is: if the 5' end is a medium mismatch (A-A, C-C, G-G, T-T), then a moderate mismatch should be introduced; if the 5' end is a strong mismatch (G-A, T-C), a weak mismatch should be introduced (G-T, A-C); if the 5' end is a weak mismatch (G-T, A-C), then a strong mismatch (G-A, T-C) will be introduced.
[0088] The present invention artificially introduces a mismatch at the 5' end of the probe, so that the connection specificity is greatly improved. Taking the R111X site as an example, no mismatch is introduced at the 5' end of the probe ( Figure 11 In a) and introducing a mismatch ( Figure 11 The difference in b).
[0089] In the process of using multiple connection detection technology to detect SNPs, the specif...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com