Liver cancer correlated leucine rich repeat and sterile alpha motif containing 1 gene methylation locus and application thereof
A technology of methylation and alpha, applied in the field of methylation determination of ribonucleic acid (ribonucleic acid) methylation sites in liver cancer, can solve problems such as no research reports.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0018] The present invention will be described in further detail below in conjunction with the accompanying drawings and specific embodiments.
[0019] 1. Analysis of heterosexual loci
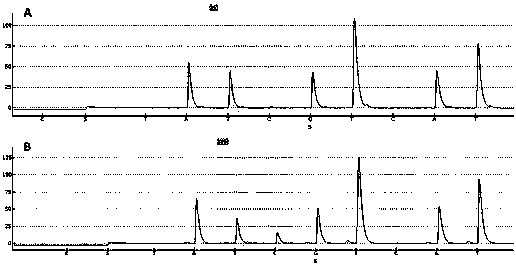
[0020] Using the Infinium HumanMethylation27 BeadChip methylation chip to screen the difference between the methylation profiles of primary hepatocellular carcinoma and corresponding paracancerous tissues, it was found that in the genomic DNA of primary hepatocellular carcinoma, the promoter region of the LRSAM1 gene The methylation level of one site (number of the site in the methylation chip: (cg05840553)) was significantly lower than that of the paracancerous group. The sequence of the specific site is (SEQ ID No.1):
[0021] GGCATGAGAGACACAGCCTGCCACATTAACTGCTAGGTTTACATCTACAACACAGCTGGAAA[CG]CTCACTGGATTCTTCACCCACACCCCGTTTAATCTCCCAACAACGCCATGCCCACTT CCTG
[0022] In order to test the accuracy of the screening results, relevant specific primers were designed, and the specificity of the specif...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com