Tumor detection kit and its special substance for recognizing n-acetylglucosamine
A technology of acetamido and kits, applied in the direction of measuring devices, recombinant DNA technology, DNA / RNA fragments, etc., can solve problems such as difficult replication and unclear structure, and achieve the effect of improving early diagnosis rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
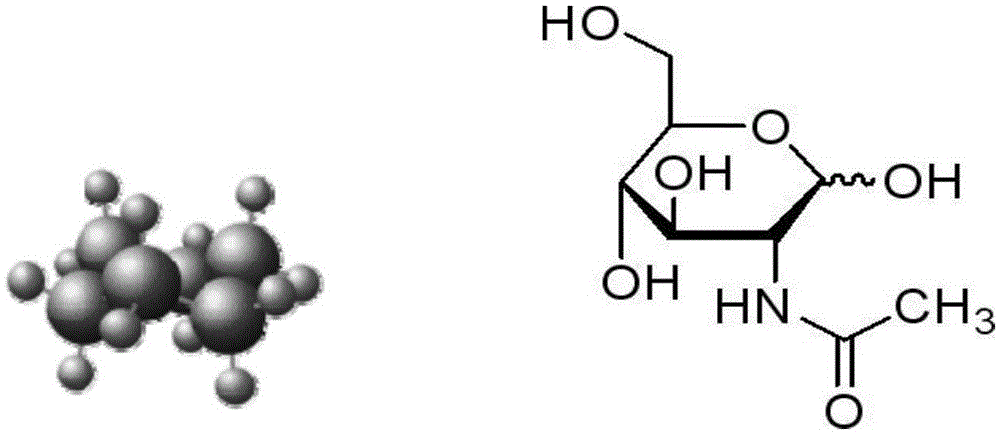
[0078] Example 1. Discovery and identification of N-acetylglucosamine for tumor detection
[0079] The inventors of the present invention have found that the content of N-acetylglucosamine in the blood of tumor patients is significantly different from that in healthy people.
[0080] In the present invention, the blood of tumor patients is directly analyzed by SELDI and MELDI mass spectrometry in vitro, to obtain the special sugar structure map of tumor patients, and to find the specific spectral lines of tumors, thereby discovering tumor-specific sugar structures, Glycoprotein, what we have discovered so far is the special sugar group of GlcNAc (N-acetylglucosamine, N-acetylglucosamine), and then the target sugar obtained by separation and purification by polyacrylamide electrophoresis, high pressure liquid chromatography, and lectin affinity chromatography protein.
[0081] We tried the method of affinity chromatography (Mu-GlcNAc was attached to the monoclonal antibody Ma6...
Embodiment 2
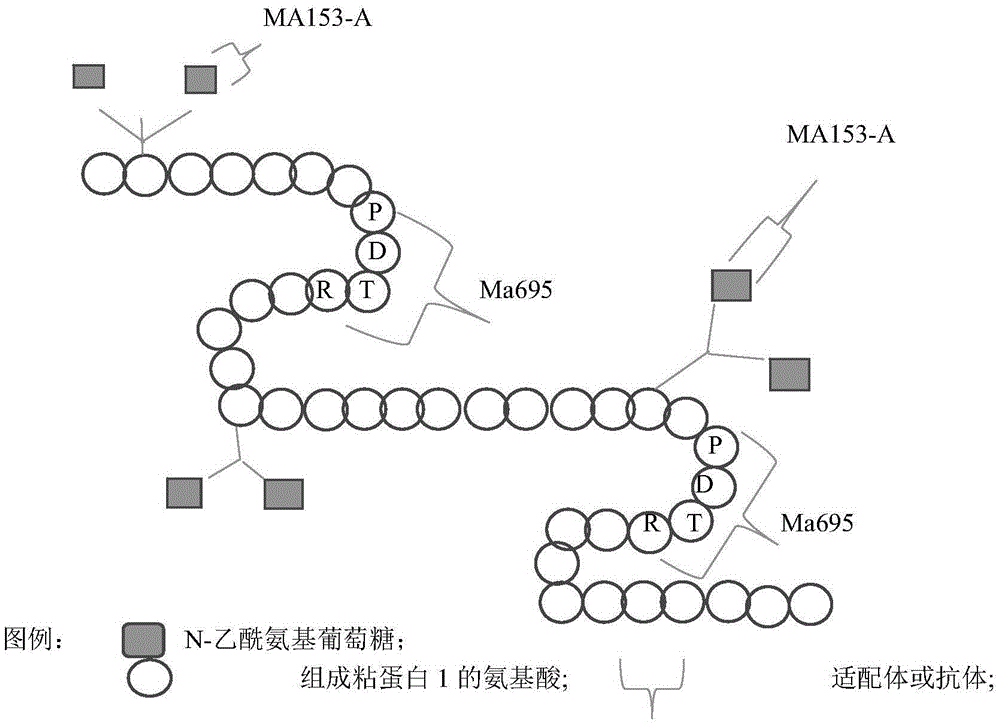
[0090] Example 2. A method for screening specific binding bodies to detect tumors using N-acetylglucosamine (sandwich method)
[0091] Determination of Mu-GlcNAc antigen in sandwich mode, the principle is as follows figure 2 shown.
[0092] (1) Materials and methods
[0093] 1 Reagents and tumor patient serum samples
[0094] 1.1 Reagent Ma695 monoclonal antibody (Mab), CanAgDiagnosticsAB, Sweden, biotin (biotin)-linked Aptamer (the aptamer shown in sequence 1 prepared as in Example 1, biotin-labeled), Mu-GlcNAc antigen, self-affinity and chromatographic extraction (see Example 1); it can also be purchased from Beijing Yonghan Xinggang Biotechnology Co., Ltd.
[0095] 1.2 Serum specimens of cancer patients The blood samples of cancer patients were obtained from the Cancer Hospital of the Chinese Academy of Medical Sciences and the 301 Hospital of the People's Liberation Army, and the diagnosis of the patients was confirmed by pathology.
[0096] 2 methods
[0097] 2.1 Pr...
Embodiment 3
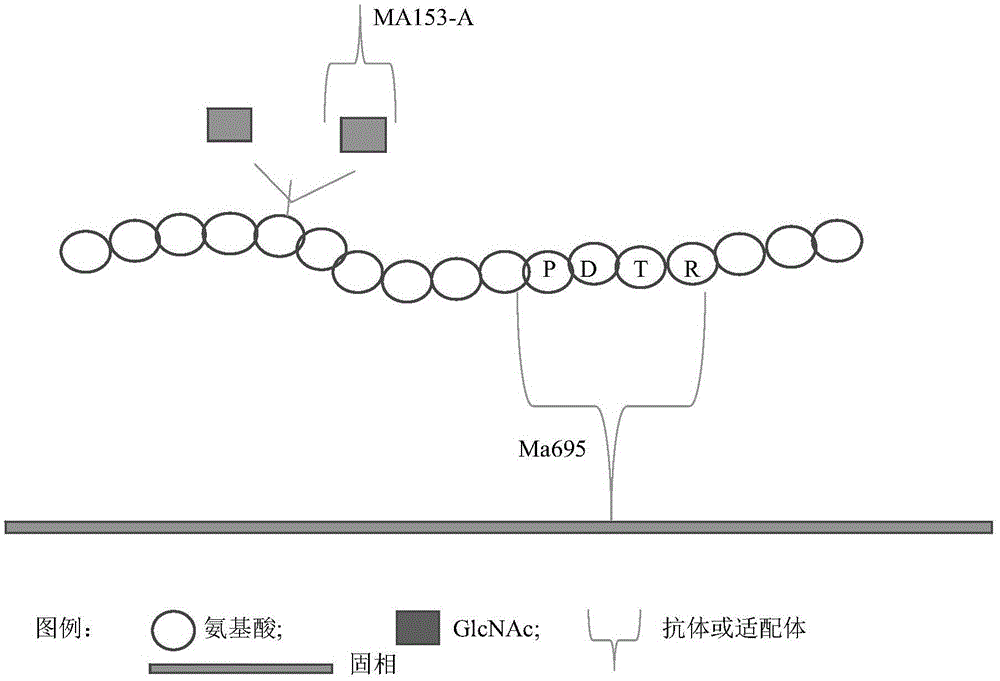
[0116] Embodiment 3, single conjugate assay method detects tumor
[0117] The principle of the single-conjugate assay method is as follows: Figure 4A and Figure 4B , using the specific binding body aptamer (shown in sequence 1) of N-acetylglucosamine. Single-conjugate analysis uses only N-acetylglucosamine-specific binding body as the recognition body; single-conjugate analysis uses N-acetylglucosamine-specific binding body to directly identify N-acetylglucosamine in the detection object; The essence of conjugate analysis is the specific combination of conjugate and -GlcNAc.
[0118] The specific method is as follows:
[0119] (1) Materials and methods:
[0120] 1 Reagents and tumor patient serum samples
[0121] 1.1 Reagents: aptamer biotin-MA153-A, Beijing Yonghan Xinggang Biotechnology Co., Ltd.; MA153 antigen, Beijing Yonghan Xinggang Biotechnology Co., Ltd.; Streptavidin, SigmaUSA.
[0122] 1.2 Serum specimens of cancer patients The serum samples of cancer patient...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com