Novel epigenetic biomarker for detecting bladder cancer and method thereof
A technology for biomarkers and bladder cancer, applied in biochemical equipment and methods, microbe determination/testing, DNA/RNA fragments, etc., can solve problems such as not good methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] The methylation array identified new methylation target genes in bladder cancer patient samples: we used the illumina infinium27K methylation array to select a higher methylation ratio score (beta value > 0.5 for methylation) and compare it to normal bladder urothelial epithelium Cells and CpG island methylated regions of seven human bladder cancer samples, in which the ZNF671 target gene showed its promoter sequence was highly methylated in bladder cancer samples and was therefore selected for further analysis.
Embodiment 2
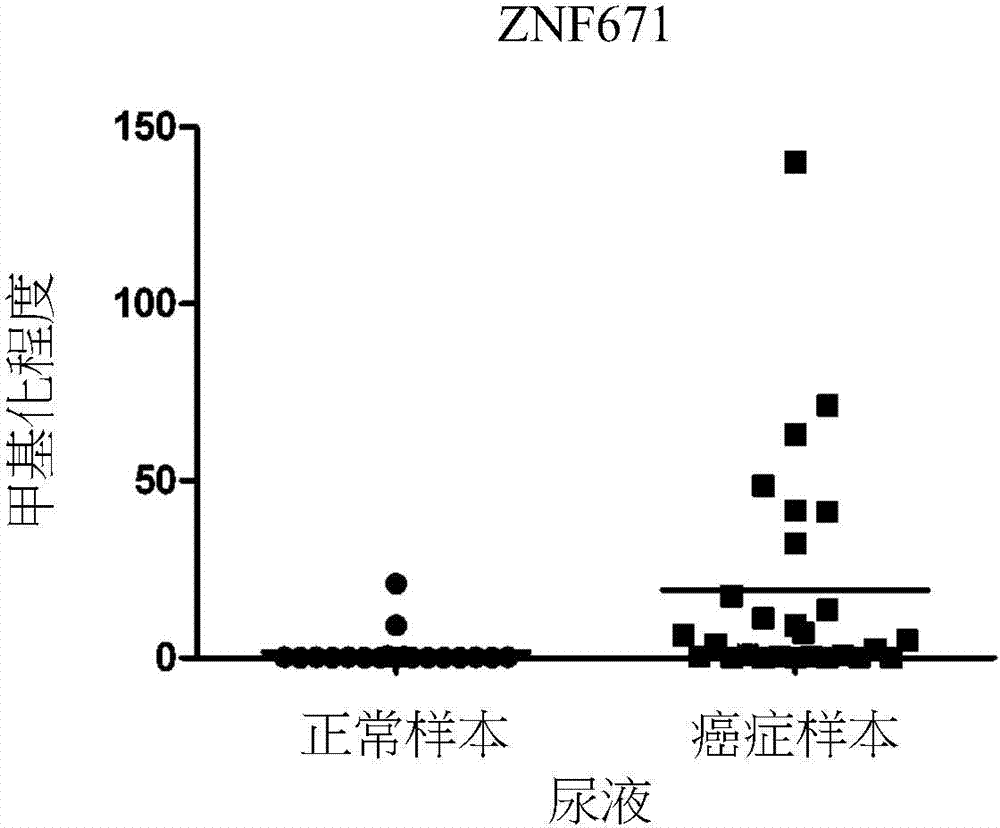
[0058] ZNF671 gene methylation in urine can be used as a detection tool:
[0059] To assess the feasibility of using DNA methylation as a marker of cancer detection and recurrence monitoring, methylation was detected using the more sensitive quantitative MSP (QMSP) method in urine from 28 bladder cancer patients and 19 non-cancer patients state( figure 1 ). The average number of ZNF671 in normal samples was 1.69, and the average number in cancer samples was 19.24. Compared with the non-cancer control group, the ZNF671 gene had a higher degree of methylation in cancer samples (P<0.001). The sensitivity and specificity of ZNF671 gene in the detection of cancer were 42.3% and 100%, respectively.
Embodiment 3
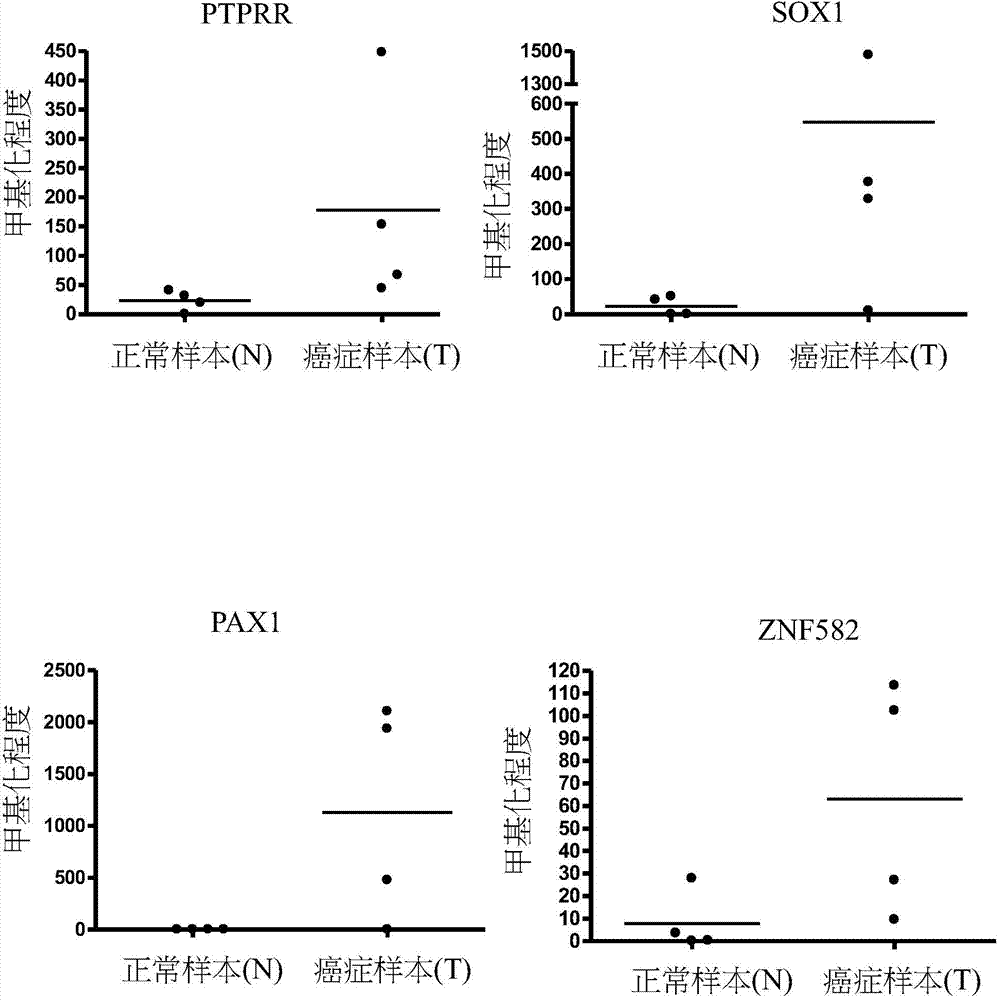
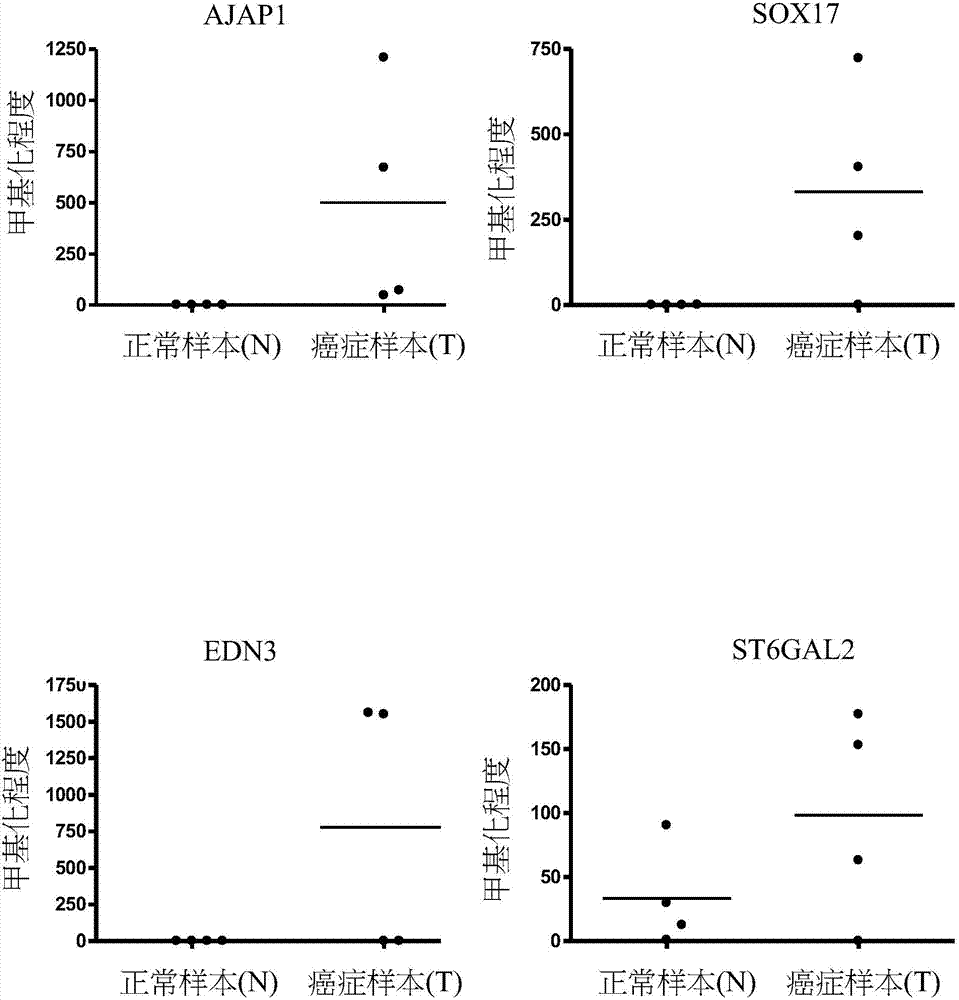
[0061] Other target gene methylation in urine can be used as a detection tool:
[0062] We also used qMSP to assess the methylation status of target genes in the urine of bladder cancer patients. The target genes are: PTPRR, SOX1, PAX1, ZNF582 ( figure 2 ); AJAP1, SOX17, EDN3, ST6GAL2 ( image 3 ); ZNF614, PTGDR, SYT9, SOX8 ( Figure 4 ); HS3ST2, POU4F3, ADRA1D, MAGI2 ( Figure 5 ); EPO, NEFH, POU4F2 ( Figure 6 ); STC2, THRB ( Figure 7 ). Table 4 shows the average number of target gene normal and bladder cancer samples in each group. The results showed that compared with normal non-cancer urine samples, these target genes had a higher degree of methylation in the urine samples of bladder cancer patients.
[0063] Table 4. The degree of methylation of each target gene
[0064]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com