Triple negative breast cancer marker COL4A2 and application thereof
A triple-negative breast cancer, marker technology, applied in the field of molecular biology and tumor drugs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Example 1 Real-time PCR detection of COL4A2 gene mRNA expression in each breast cancer cell
[0020] 1. Experimental materials:
[0021] Individual cells and their media:
[0022] (1)MDA-MB-231: L15+10%FBS
[0023] (2) MCF-7: DMEM-H+10%FBS+0.01mg / ml insulin
[0024] (3) BT-474: RPMI1640++10%FBS+0.01mg / ml insulin
[0025] (4)SK-BR-3: RPMI1640++10%FBS
[0026] 2. Experimental method:
[0027] 2.1 Extraction of total cellular RNA:
[0028] Collect about 1 × 10 cultured cells after transfection 6 One, wash twice with PBS; add 1ml of pre-cooled TRNzol to the centrifuge tube, mix well, place at room temperature for 10min, centrifuge at 10000rpm for 10min at 4°C, take the supernatant and transfer to a new centrifuge tube; add 0.2ml of chloroform , cover the tube cap, shake vigorously for 15 seconds, and place at room temperature for 3 minutes; centrifuge at 10,000 rpm for 10 minutes at 4°C, take the upper colorless aqueous phase, transfer to a new centrifuge tube, add a...
Embodiment 2
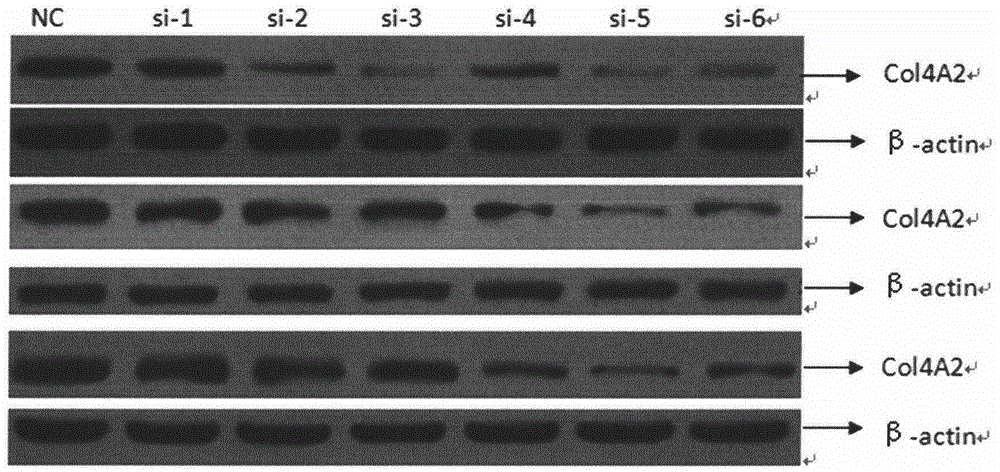
[0045] Example 2 Western blot detection of COL4A2 protein expression in each breast cancer cell
[0046] 1. Experimental method
[0047] 1.1 Extraction of target protein:
[0048]After the cell culture was completed, the cell culture medium in the culture plate was discarded, washed twice with 0.9% NaCl pre-cooled at 4°C, the NaCl was blotted dry, and the cell culture plate was placed on ice. Add 100 μl of RIPA cell lysate pre-added with protease inhibitors to each well, and lyse the cells on ice for 3 min. Scrape the lysed cells with a cell scraper, transfer the cell lysate to a pre-cooled 1.5ml EP tube on ice, and centrifuge at 12000rpm at 4°C for 20min. After transfer and centrifugation, transfer the supernatant to a new pre-cooled 1.5ml EP tube and measure the protein concentration.
[0049] 1.2 Determination of protein concentration:
[0050] According to the number of standards and samples, prepare an appropriate amount of BCA working solution with 50 volumes of BCA ...
Embodiment 3
[0073] Construction of embodiment 3 interference plasmid
[0074] 3.1 Design and synthesis of COL4A2 gene interference sequence
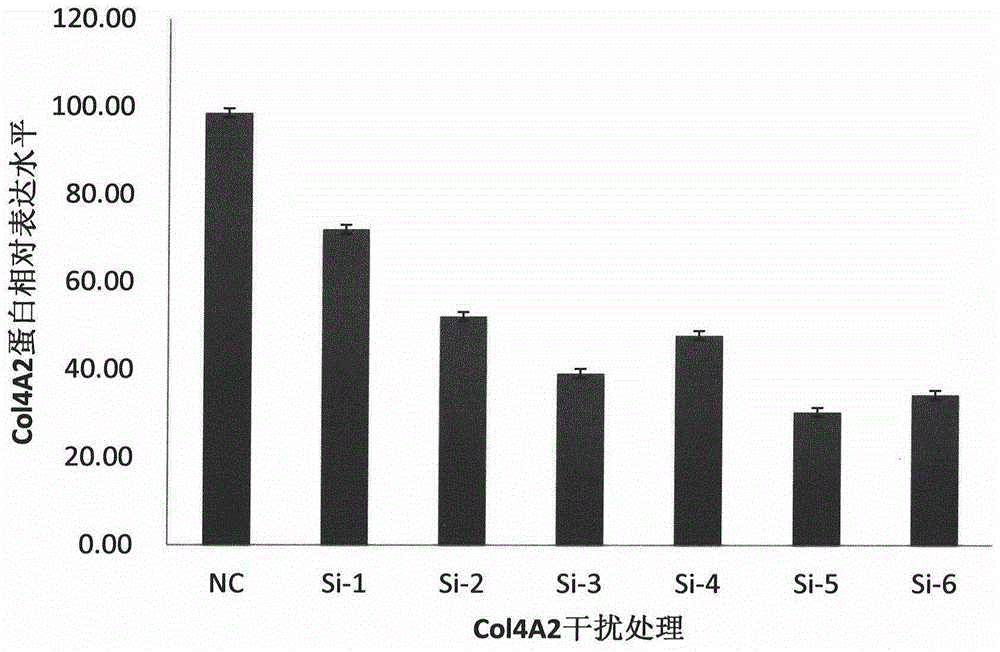
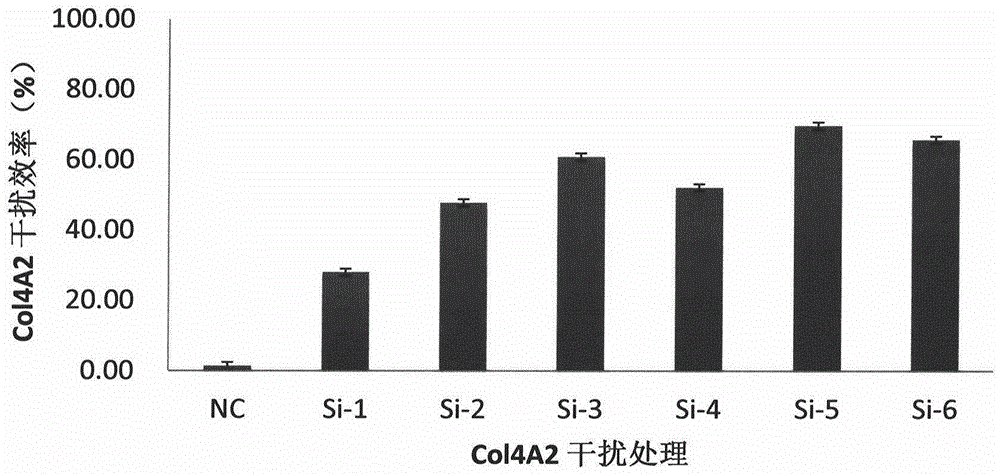
[0075] According to the gene sequence of COL4A2 (Gene ID: 1284) in GenBank, 6 interference sequences and 1 negative control sequence were designed. The interference sequences and their nomenclature are shown in Table 8:
[0076] Table 8 Design of interference sequences
[0077] serial number
sequence naming
sequence
SEQ ID NO 1
Si-1
GGGTGTGAAGAAGTTTGAT
SEQ ID NO 2
Si-2
GCCTTATGCACTGCCTAAA
SEQ ID NO 3
Si-3
GGGGTGAACCTGGAGAGCC
SEQ ID NO 4
Si-4
GCCAAGACCAAGGAGAATG
SEQ ID NO 5
Si-5
GGAATGCAGATGTACAGAA
SEQ ID NO 6
Si-6
GGCAACAGAGGACTTGGTT
SEQ ID NO 7
Neg
GCAGATAGGTAGGCGTTAT
[0078] 3.2 COL4A2 interference sequence primer design
[0079] The primer has a HindIII restriction site at the 5' end and a BamHI restriction site at t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com