Bi-order genetic calculation-based gene expression data bi-clustering algorithm
A gene expression and biclustering technology, applied in the field of data mining processing, can solve the problems of large data volume, high dimensionality, high redundancy, etc., and achieve the effect of wide search range, high quality, and overcoming local information.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0027] The present invention will be further described in detail below in conjunction with the embodiments and the accompanying drawings, but the embodiments of the present invention are not limited thereto.
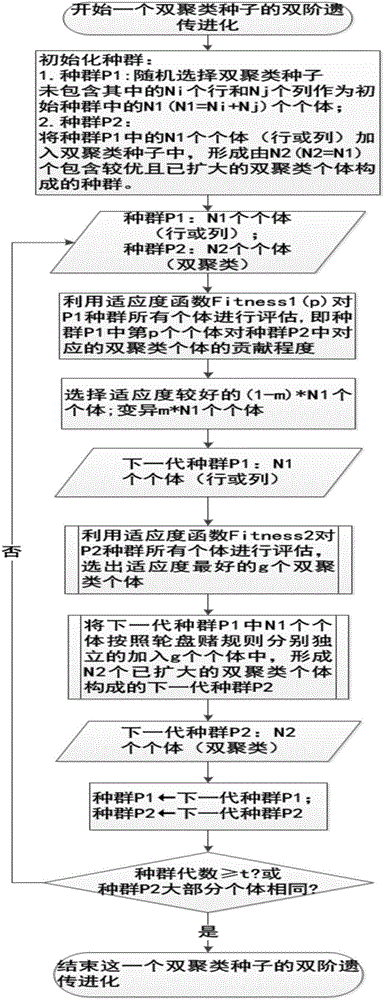
[0028] like figure 1 , a biclustering algorithm based on two-stage genetic computation, includes steps in the following order:
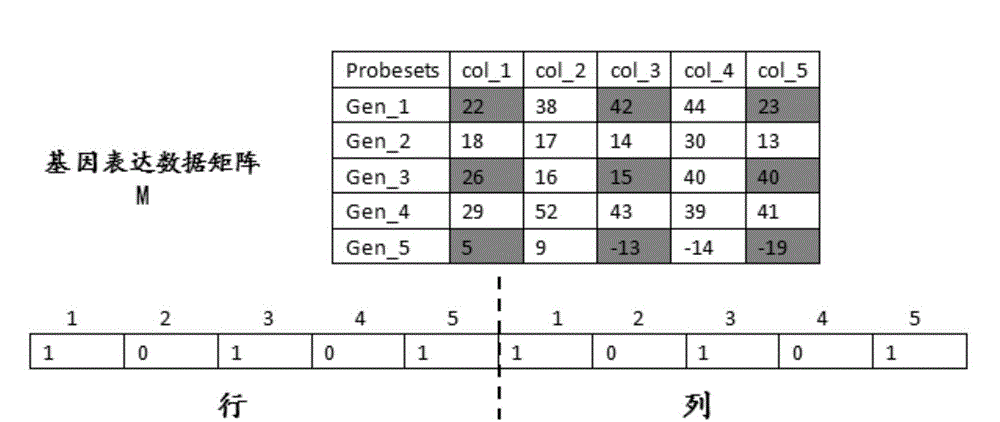
[0029] 1) The gene expression data matrix is M, the number of rows is m, and the number of columns is n, that is, the size of the gene expression data matrix is m×n. Subtract the k-th row from each row of the original data matrix M to obtain the processed Matrix M(k), k=1,2,...,n;
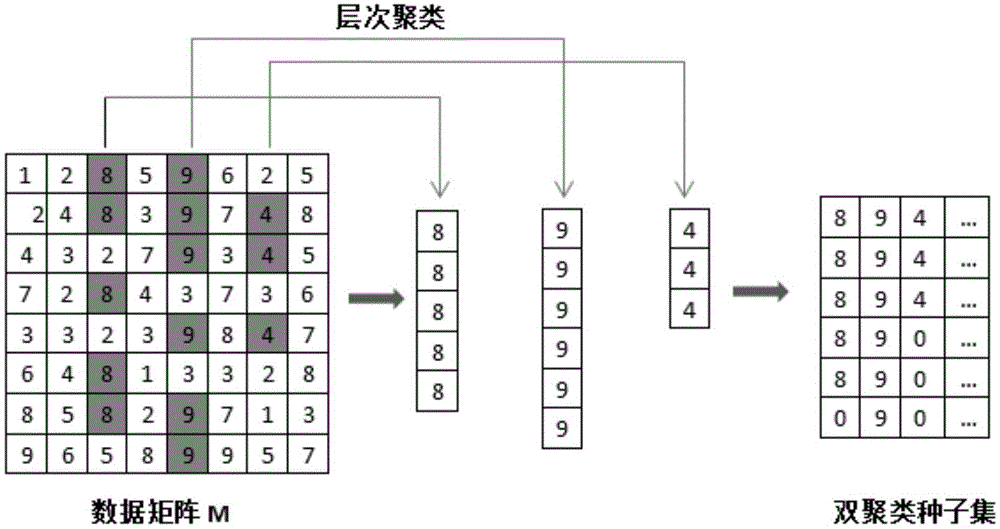
[0030] 2) For each column except the kth column in the processed matrix M(k), such as figure 2 As shown, use hierarchical clustering with a distance threshold of cof=0.02 for each column to obtain the biclustering seeds of each column, and then put all the obtained biclustering seeds into a set named Bic_Set; because each The bi-clustering seeds correspond t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com