Non-invasive determination of methylome of fetus or tumor from plasma
A methylation, group region technology, applied in the field of methylation pattern determination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0069] definition
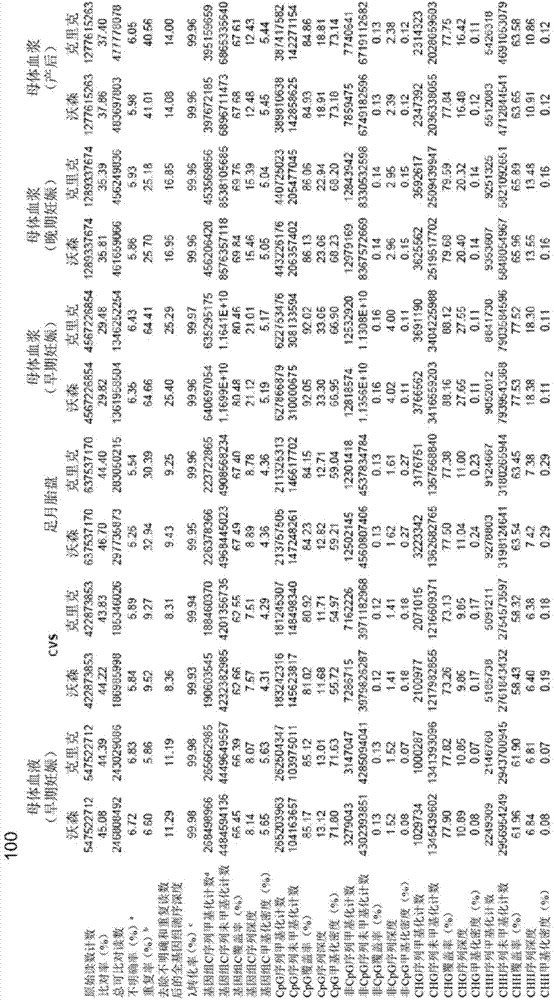
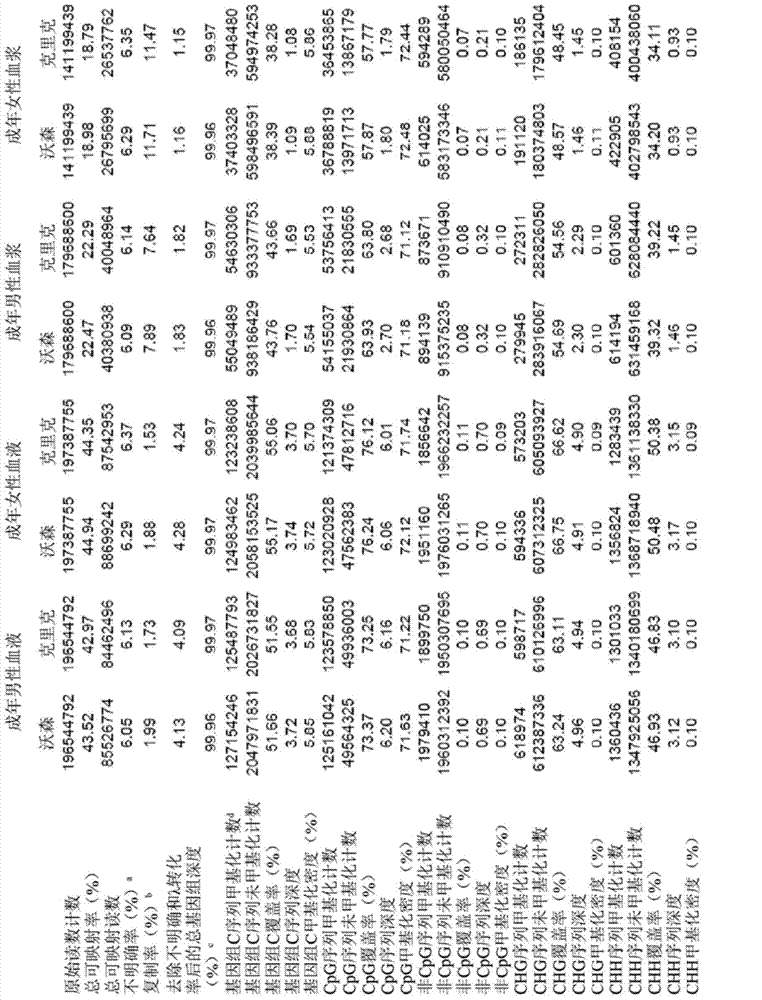
[0070] A "methylome" provides a measure of the amount of DNA methylation at various sites or loci in a genome. A methylome may correspond to all of the genome, a large portion of the genome, or a relatively small portion of the genome. A "fetal methylome" corresponds to the methylome of a fetus of a pregnant female. The fetal methylome can be determined using a variety of fetal tissues or sources of fetal DNA, including placental tissue and cell-free fetal DNA in maternal plasma. A "tumor methylome" corresponds to the methylome of a tumor of an organism, such as a human. Tumor methylomes can be determined using cell-free tumor DNA in tumor tissue or maternal plasma. Fetal methylomes and tumor methylomes are examples of related methylomes. Other examples of relevant methylomes are those that can contribute DNA to bodily fluids (e.g. plasma, serum, sweat, saliva, urine, genital secretions, semen, fecal fluids, diarrheal fluids, cerebrospinal fluid, gastro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com