Hepatoma cell-specific aptamer and preparation method thereof
A liver cancer cell and aptamer technology, applied in the field of genetic engineering, can solve the problems of low binding force or specificity of liver cancer cells, achieve sensitive early diagnosis methods, and improve the effect of diagnosis and treatment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
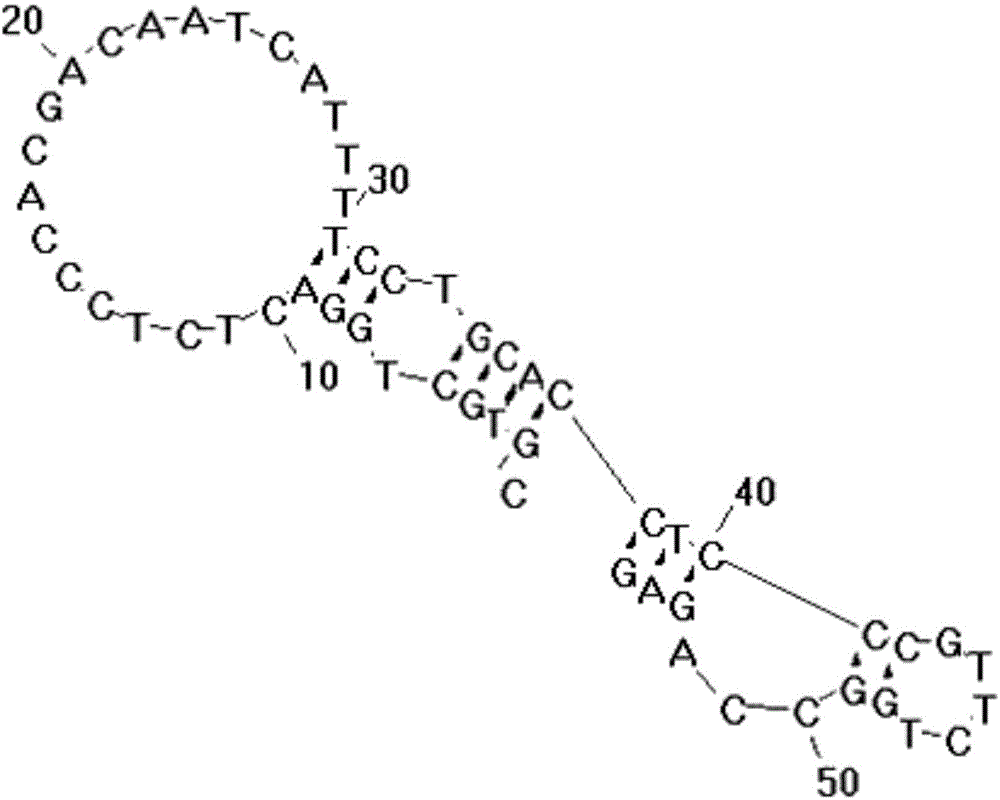
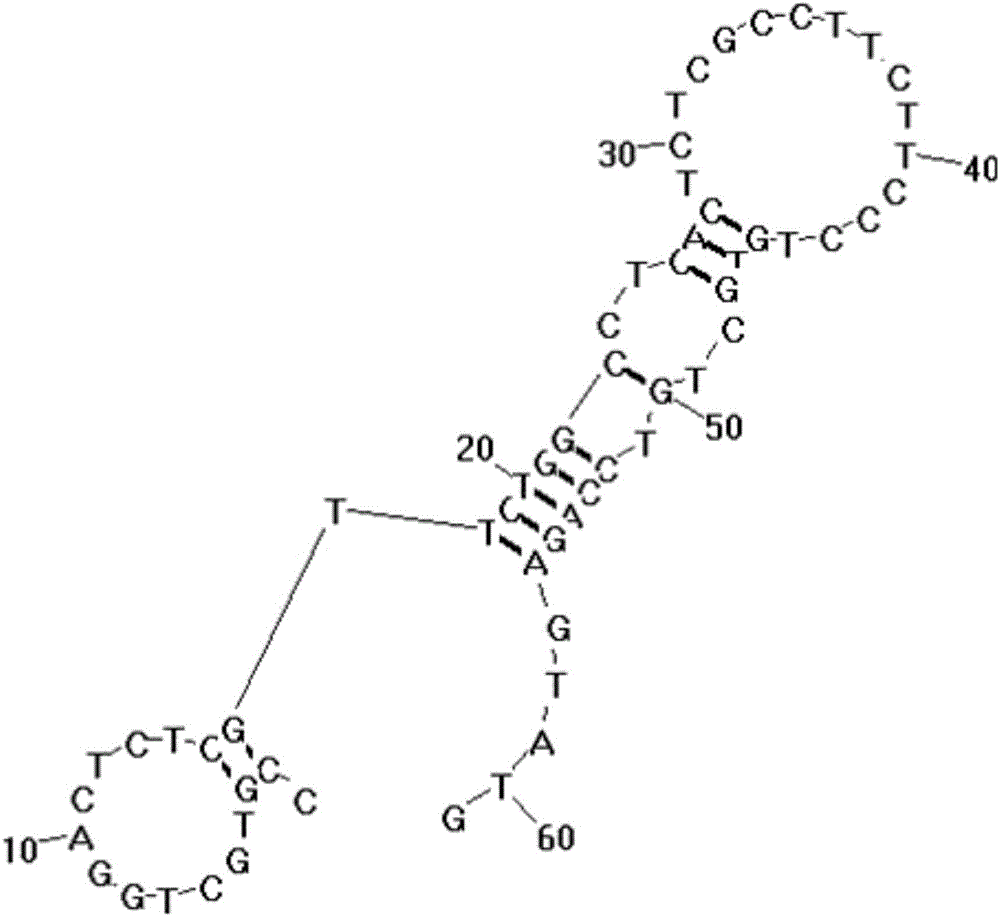
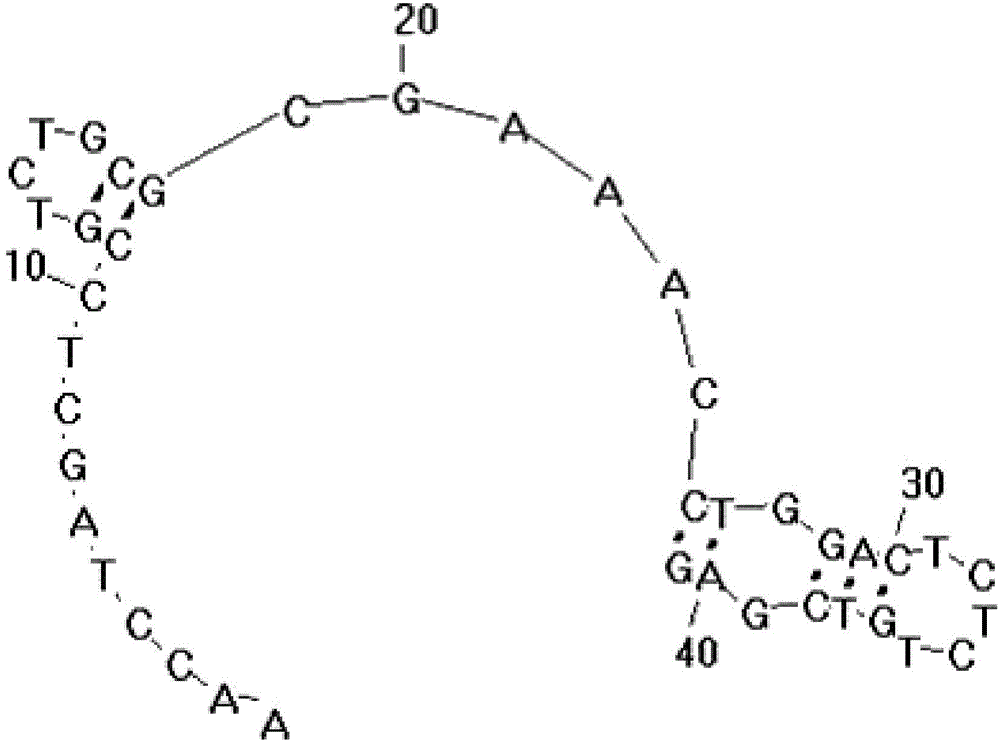
[0050] The embodiment of the present invention provides a liver cancer cell-specific aptamer, which may include: the H2c aptamer shown in the sequence listing SEQ ID NO: 1, the H2c aptamer shown in the sequence listing SEQ ID NO: 2 At least one of the H4c adapter and the H5b adapter shown in the sequence table SEQ ID NO: 3, wherein the structures of the three adapters are as follows Figure 1-3 shown.
[0051] Specifically, the 5' end of the aptamer can be connected with the fluorescent group FAM, Cy5 or Cy3, and the 3' end of the aptamer can be connected with the quenching group BHQ1 or BHQ2. Among them, when the above three aptamers are used for fluorescence experiments, the 5' end can be connected to the fluorescent group Cy5, and when the above-mentioned aptamers are used as fluorescent probes, the 5' end can be connected to the fluorescent group Cy5 or Cy3 .
[0052] The aptamer provided in the embodiment of the present invention can recognize liver cancer cells with hi...
Embodiment 2
[0054]The embodiment of the present invention provides a method for preparing the aptamer provided by the first embodiment of the present invention, such as Figure 21 As shown, the method includes:
[0055] Step 1: Synthesizing the first library of random single-stranded oligonucleotides and primers, wherein the first library of single-stranded oligonucleotides is 5′-ACCGACCGTGCTGGACTCT-N 40 - AGTATGAGCGAGCGTTGCG-3', the primers include a forward primer and a reverse primer, the forward primer is shown in SEQ ID NO: 4 in the sequence listing, and the reverse primer is shown in SEQ ID NO: 5 in the sequence listing.
[0056] Step 2: Carry out forward screening, which includes: co-incubating the first library with target cells, wherein the incubation temperature is 37°C, and the target cells can be at least one of Huh7.5.1 liver cancer cells and HepG2 liver cancer cells , the target cells selected in this implementation are Huh7.5.1 liver cancer cells.
[0057] Step 3: Separat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com