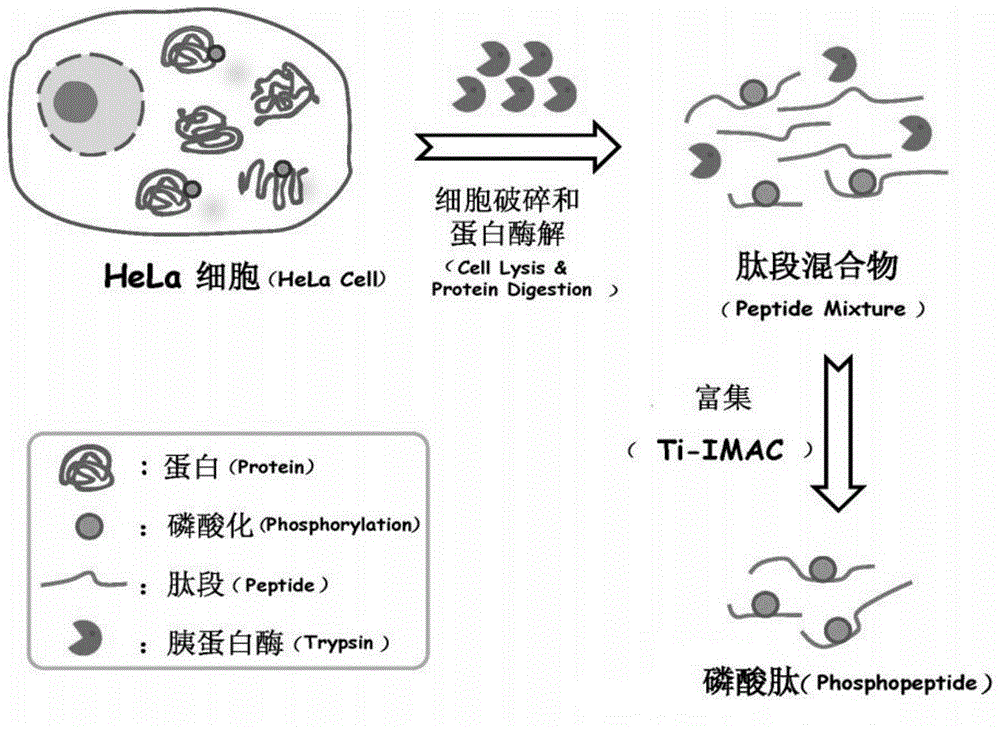
Rapid treatment method for phosphoproteome sample
A technology for phosphorylating proteins and processing methods, applied in the preparation of test samples, etc., can solve the problems of high non-specific adsorption performance of immobilized enzymes, no generalization, inconvenient operation, etc., and achieve high-quality spectral identification coverage. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
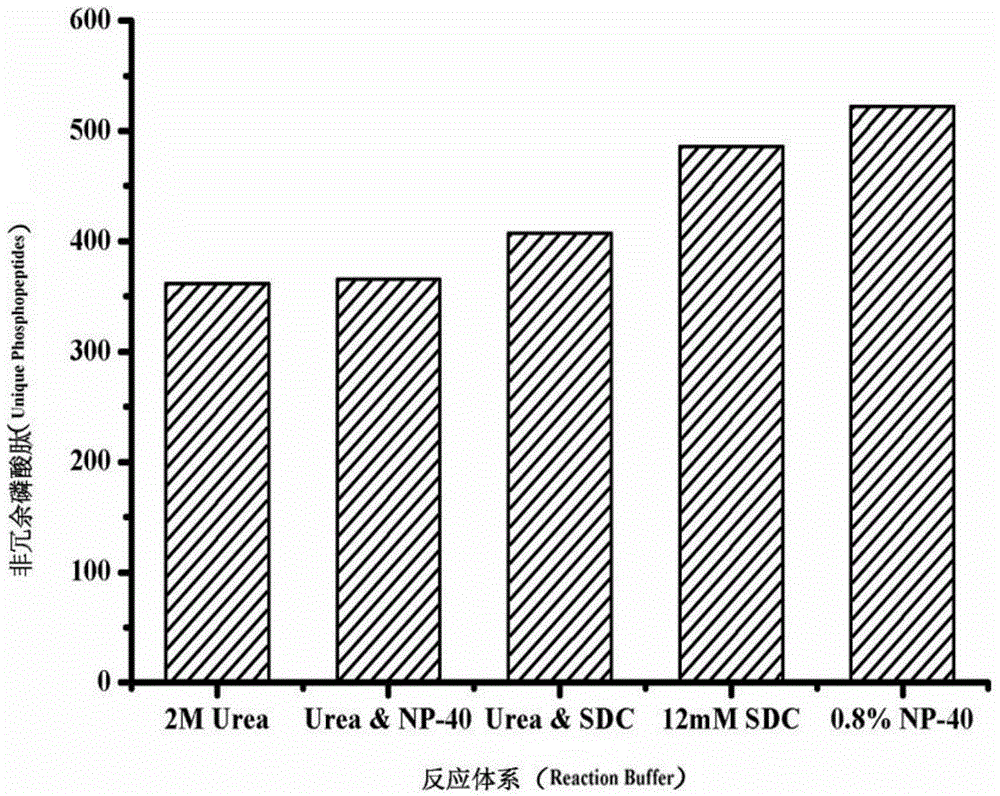
[0030] Investigation of the reaction system of cell lysis-proteolysis: 100,000 HeLa cell samples were placed in the following five reaction systems, namely: 1) 2M urea buffer solution; 2) 0.8% NP-40 buffer solution; 3) 12mM sodium deoxycholate (SDC) buffer solution; 4) 2M urea and 0.8% NP-40 buffer solution; 5) 2M urea and 12mM SDC buffer solution, these five buffer solutions contain 10mM DTT and phosphatase inhibitor respectively (ie 1mM NaF and 1mM Na3VO4 ), then add 300ng / μL trypsin to the solution, vortex and shake on the shaker for tens of seconds until all the cell membranes are broken, then place the mixture in a 37°C water bath sonicator for ultrasonic incubation for 1h, and finally, control it by centrifugal force A GELoader Tip centrifuge filled with 1 mg Ti-IMAC material was used for enrichment of phosphopeptides. The obtained phosphopeptides were lyophilized at room temperature and redissolved in 100 μL 0.1% (v / v) formic acid for RP LC-MS / MS For analysis, qualitati...
Embodiment 2
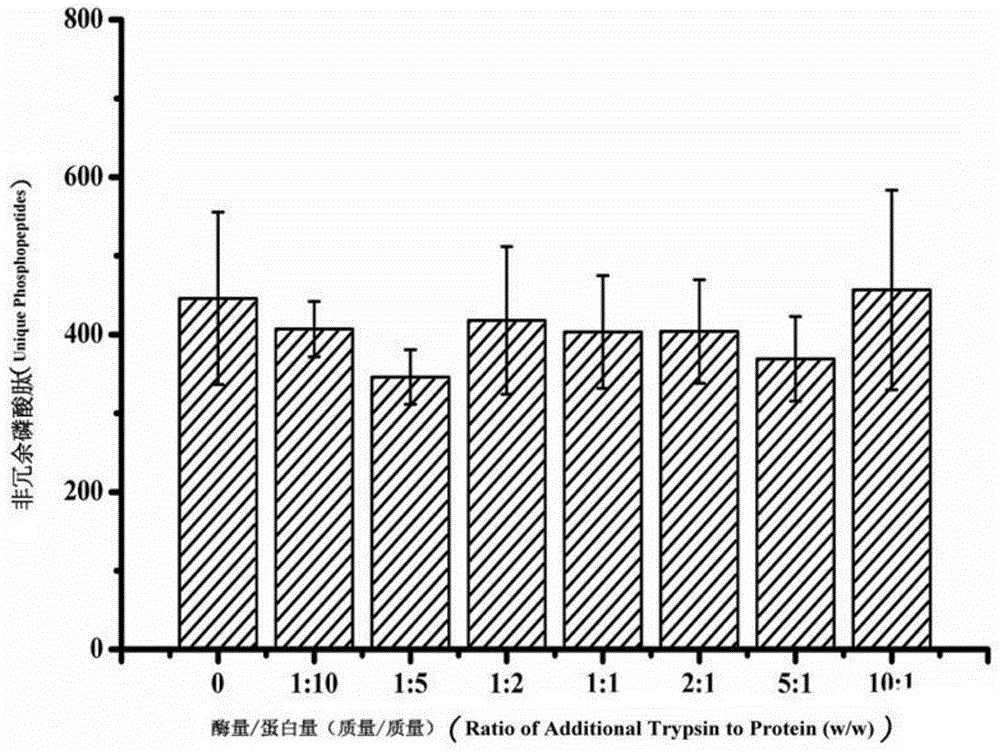
[0033] The effect of high-concentration trypsin on the enrichment of phosphopeptides and identification by mass spectrometry: Add a series of trypsin with different enzyme / protein ratios to 200 μg of HeLa hydrolyzate, mix well, and enrich with 2 mg of Ti-IMAC material. The enrichment steps of phosphopeptides are as follows: After resuspending the material with 80% ACN6%TFA, mix it with the enzymatic hydrolysis solution at a volume ratio of 1:1, shake at room temperature for 30 minutes, and after fully enriched, at 13500rpm Centrifuge for 3 minutes, discard the supernatant, then wash with 50% ACN6%TFA, 200mM NaCl and 30% ACN6%TFA in sequence, shake for 30 minutes, remove the supernatant by high-speed centrifugation as above, and finally wash with 10% NH 3 ·H 2 The phosphopeptide was eluted twice with O solution and subsequently identified by mass spectrometry as described in Example 1.
[0034] Such as image 3 As shown, the numbers on the axis of abscissa represent the exces...
Embodiment 3
[0036] Optimization of the reaction conditions for the rapid processing method of phosphorylated proteome samples: the optimal trypsin concentration and reaction time required for the reaction were investigated respectively. Add four groups of 100000 HeLa cell samples into 50 μL 0.8% NP-40 reaction system, then add 20ng / μL, 100ng / μL, 300ng / μL, 1.0μg / μL trypsin respectively, and vortex the solution on the shaker Rotate and oscillate for tens of seconds until all the cell membranes are broken, then place the mixture solution in a 37°C water bath ultrasonicator and incubate for 1 hour, and finally, use a GELoader Tip centrifuge with controllable centrifugal force and filled with 1mg Ti-IMAC material for phosphoric acid Peptide enrichment, as described in Example 1, was followed by mass spectrometric identification.
[0037] As mentioned above, after the optimal concentration of trypsin was investigated, four groups of 100,000 HeLa cell samples were added to the reaction system of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com